Abstract
Glioblastoma multiforme (GBM) was shown to harbor therapy-resistant cancer stem cells that were major causes of recurrence. PDGFR (platelet-derived growth factor receptor) and c-Kit (stem cell factor receptor) signaling play important roles in initiation and maintenance of malignant glioma. This study demonstrated that long-term culture with imatinib mesylate, the tyrosine kinase inhibitor against PDGFR and c-Kit resulted in reduced cancer stem cell ability in glioblastoma cells through cell differentiation. Derived from RG glioblastoma cells co-cultured with imatinib for 3 months, RG-IM cells showed distinct properties of cell cycle distribution and morphology in addition to significantly decreased ability to form aggregates and colonies in vitro and tumorigenicity in vivo. Increased expression of GFAP (astrocyte marker) and class III β-tubulin isotype (Tuj1, neuron marker) were detected with morphology like neurons or astrocytes in RG-IM cells. Furthermore, decreased expression of stem cell markers, i.e., CD133, Oct-3/4, nestin, and Bmi1, and increased terminal neural cell markers, GFAP, Tuj1, etc., were identified in RG-IM at the mRNA level. All these markers were changed in RG cells when PDGFRB and c-Kit expression were double knocked down by siRNA. Cell differentiation agent, all-trans retinoic acid (ATRA) caused similar effect as that with imatinib in RG cells, while adding PDGF-B and SCF in RG-IM resulted in cell dedifferentiation to some extent. Moreover, differentiation in RG cells treated by imatinib or ATRA was mainly driven by MAPK signaling pathways. In summary, continuous inhibition on PDGFR and c-Kit signaling disturbed glioma stem cells biology in subsets of GBM cells and may have potentials in clinical applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma Multiforme (GBM, WHO grade IV astrocytoma) is the most common and aggressive brain tumor in adults and has the poorest clinical prognosis despite significant advances in cancer therapy. Recent studies suggest that GBM contains cellular subpopulations with potent tumorigenesis and a few stem cell characteristics [1]. These glioma stem cells (GSC) express the neural precursor cell surface marker CD133 (prominin-1), self-renew as displayed by serial neurosphere formation, and contribute not only to tumor initiation and maintenance but also to its therapeutic resistance [1]. Thus far, however, whether these GSC derive from malignant transformation of neural progenitors or dedifferentiation of mature astrocytes during gliomagenesis is largely unknown [2]. GBM is a complex tumor with multiple genetic alterations and aberrant cellular signaling pathways, it is likely that the GSC may co-opt with these alterations, such as mitogenic cues to maintain their stem cell properties.
Constitutive activation of growth factor receptors and their downstream signaling pathways is a hallmark of malignant glioma. Among them, the platelet-derived growth factor receptor (PDGFR) and their cognate ligands, PDGF-A and PDGF-B are over-expressed in human glioma cell lines and gliomas with higher level of expression correlating with increasing tumor grade and a poorer prognosis [3, 4]. This suggests that both autocrine and paracrine stimulation could play an important role in initiation and maintenance of the tumor. PDGF is a potent growth and differentiation factor during normal development [5]. A causal relationship between the PDGF signaling and the development of gliomas is marked by the fact that retroviral delivery of PDGF-B in the brain of neonatal mice results in the formation of brain tumors [6]. Introduction of PDGF to astrocytes or nestin-expressing progenitors led to glioma formation, a higher grade could be achieved when combined with Ink4a-Arf deficiency [7]. Stem cell factor receptor (c-Kit) and its ligand stem cell factor (SCF) are known to have significant roles in gametogenesis and hematopoiesis [8]. SCF/c-Kit signalings are also involved in nervous system development. Moreover, as activated in many types of cancers, c-Kit is found either over-expressed, amplified, or both in glioma [9]. Interestingly, c-Kit is located at chromosome locus 4q12 that contains two other receptor tyrosine kinase genes, PDGFRA, and VEGFR2 [8]. Co-amplification of c-Kit, PDGFRA, and VEGFR2 is a repeated finding in primary glioblastoma [9]. These evidences prompted the notion that targeted inhibition of PDGF receptors and c-Kit might directly disturb GBM biology by interrupting the growth cycle or other key features of the tumor [10].
Imatinib mesylate (imatinib, Gleevec, or STI-571; Novartis, Basel, Switzerland) is a phenylaminopyrimidine derivative which selectively inhibits the activation of several receptor tyrosine kinases, including the PDGFR, c-Kit, and the Abl tyrosine kinase [11]. Imatinib has shown significant effects not only in patients with chronic myeloid leukemia (CML) bearing Bcr-Abl activation but also proven to inhibit c-Kit- and PDGFRA-mediated signaling and improve survival in patients with gastrointestinal stromal cell tumors [12] and with hypereosinophilic syndrome [13]. However, significant anti-tumor effect of imatinib treatment has not obtained in human malignant glioma, which often expressed PDGFR and/or c-Kit [14]. Heterogeneity of glioblastoma and non-tailored treatment regimens might account for such results [2]. We previously demonstrated that imatinib treatment resulted in differential effect in various human glioblastoma cells, including U87MG, T98G, RG, etc. [15]. In addition, the treatment stimulated significant activation of ERK (p44/42 MAPK), which paralleled with diverse cellular effects in different PDGFR-expressing glioblastoma cells [15]. In this study, we demonstrated that continuous treatment with imatinib significantly disturbed glioma stem cell biology in a GBM-derived cell line. Our results indicated that PDGFR and/or c-Kit signaling may contribute to the maintenance of undifferentiated cells in glioblastoma, and targeted inhibition on the related cell signaling may decrease the malignant potential of the tumor. Our data may thus provide considerable information on developing tailored treatment regimens for patients with malignant glioma.
Materials and methods
Drugs and reagents
Imatinib mesylate (STI571, Gleevec®) was obtained from Novartis Co. Ltd. as described before [15]. All-trans retinoic acid (ATRA) was stocked in DMSO to the concentration of 3 mg/ml. Human recombinant SCF and PDGF-B (Peprotech EC, London, UK) were dissolved in PBS or H2O to a concentration of 100 μg/ml. The MEK1/2 inhibitor U0126, the PI3 K inhibitor LY294002, and PI3K/mTOR inhibitor PI103 were dissolved in DMSO and stored at −20 °C. Drugs were further diluted in cell culture medium to their respective final concentrations when investigated for their effect in cell culture. The protein kinase inhibitors and reagents were purchased from Sigma-Aldrich, Shanghai, China, unless specified.
Glioblastoma cell lines
RG glioblastoma cells were originally primary tissue cultures from tumor biopsy [16]. The imatinib-resistant RG cell line (RG-IM) was gradually established by continuously co-culturing with 10 μM of imatinib for up to 3 months. The GBM cells were maintained in DMEM (Sigma) supplemented with 10 % heat-inactivated fetal calf serum (Sigma), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified incubator with 5 % CO2 and serially passaged every 2–3 days.
Tumor sphere culture in vitro
Hanging-drop method was used to culture tumor cell spheres in vitro [17]. Briefly, drops of the cell suspension (20 μl) containing 3,000–7,000 tumor cells were placed onto the inner face of the lid of a 100-mm petri-dish, the lid was then inverted to cover over the dish containing 10 ml DMEM. Hanging-drop cultures were incubated for a period of 3 days. The resulting cell aggregates or spheres were harvested using a 1,000 μl Pasteur pipette and introduced into a 24-well plate base coated with 1 % agar and filled with 1 ml of DMEM plus 10 % FCS. Establishment of the tumor sphere was checked and recorded the next 2–3 days by an inverted phase-contrast light microscope (Nikon, ELWD 0.3, Japan).
Western blotting
Bradford assays (Bio-Rad, Herts, UK) were performed to determine the total protein concentration of the whole cell lysates in Cell Lysis Buffer. The protein concentration was normalized to 1 μg/μl for all samples including the controls. Up to 30 μl of the protein lysates in sample buffer was loaded within each well. Primary antibodies directed against nestin (Abcam, Shanghai, China), total- and phospho-PDGFRB, total- and phospho-c-Kit, total- and phospho-p44/42 MAPK, total- and phospho-AKT (Ser308), total- and phospho-rpS6 and β-actin, and secondary antibodies (anti-rabbit/mouse HRP-conjugated) were applied. Enhanced chemiluminescence (Pierce Ltd., Shanghai) detection was then used and the levels of proteins and phospho-proteins were quantified by densitometry using NIH-ImageJ®. The ratio of the mean density of phospho- to that of total-protein/-actin was calculated and recorded as the relative expression level of the protein activity. The antibodies and reagents were purchased from Cell Signaling Technology Inc., Danvers, MA, if not mentioned otherwise.
Immunofluorescent microscopy
2 × 104 cells were plated on 12-well plates pre-treated with polylysine. At the harvest, cells were fixed with 4 % formaldehyde before permeabilized with 1 % Triton X-100. After blocking in solution containing 4 % goat serum and 1 % bovine serum albumin for 1 h, the cells were incubated with either mouse anti-human GFAP (1:200, Abcam, Shanghai) or mouse anti-human Class III β-Tubulin (1:200, Sigma), or mouse anti-human nestin (1:200, Abcam) for 1 h. After washing 3 times with TBST, the secondary antibody of FITC-labeled anti-mouse IgG (1:32, Sigma) was applied for 1 h in the dark. Cells were then incubated with the nuclear staining dye 4,6-diamidino-2-phenylindole (DAPI, Roche, Basel, Swiss). The images were viewed using a fluorescent microscope (Nikon ECLIPSE 80i) and captured using a CCD camera (Nikon DS-Ri1, Tokyo, Japan). All images were at 200× magnification.
Cell cycle analysis by flow cytometry
Cell cycle analysis was performed using Cycle Test™ Plus DNA Reagent Kit (BD Biosciences, Oxford, UK) on a FACStrak Flow cytometer equipped by means of CellQuest software. RG glioblastoma cells treated with 10 μM of imatinib for 48 h, RG-IM cells, and non-treated controls were prepared and harvested. Propidium iodide (PI) (Sigma) staining was carried out, flow-cytometric analysis was performed, and the cell cycle distributions were analyzed by Modfit LT™ 2.0 software (Verity Software House, Cambridge, MA).
Real-time PCR analysis
Total RNA was extracted from cultured glioblstoma cells using Trizol-Reagent (Invitrogen Ltd., Shanghai, China) and cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystem, Foster City, CA). Gene expression analyses were performed using the Power SYBR Green PCR Master Mix (Applied Biosystem). The primer sequences, the accession numbers, and the product sizes are listed in Table 1. The PCR conditions for all genes were as follows: 95 °C initial activation for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s, and fluorescence determination at the melting temperature of the product for 20 s using an ABI Step One (Applied Biosystem). The relative expression at the mRNA level was calculated and the values were normalized as fold changes in relation to the expression of house-keeping genes GAPDH and β-actin.
Small interfering RNA (siRNA) treatment
For specific gene knockdown effect on PDGFRB and/or c-Kit at the mRNA level, verified siRNA against PDGFRB, c-Kit, and the control siRNA were ordered from Invitrogen Ltd. siRNA against PDGFRB, c-Kit, PDGFRB plus c-Kit were transfected into RG cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Cells transfected with the transfection agent, but no siRNA (Mock) or scramble-control siRNA (Negative control), were used as the controls. Total RNA was prepared from the samples and controls collected at 48-h post transfection and used for real-time quantitative PCR analysis.
Colony formation assays
To assess the clonogenic ability in vitro, RG and RG-IM cells were plated in 6-well plates in appropriate densities. Cells were incubated for up to 2 weeks at 37 °C with 5 % CO2 and the cell medium containing 10 % heat-inactivated fetal calf serum (Sigma), 100 U/ml penicillin and 100 μg/ml streptomycin was refreshed every 3 days. At the harvest, colonies were fixed with 2 % ethanol and stained with 0.5 % crystal violet. A colony that contained more than 500 cells was counted. Each experiment was carried out in duplicates. Each set of the experiments was done at least 3 times. Photos of the 6-well plates at the end of cell culture were taken using a digital camera (Nikon, F500, Japan).
Tumor xenografts
For tumorigenicity experiments in vivo with subcutaneously growing human xenografts, nude mice (BALB/c, nu/nu, 6–8 weeks, 20 g) were obtained from Charles River Laboratories (Beijing, China). All mice were fed and maintained in specified pathogen-free conditions according to the guidelines of Care and Use of Laboratory Animals published by the China National Institute of Health. Human glioblastoma cells RG or RG-IM were injected subcutaneously (s.c.) into the right and left hind limb (1 × 107 cells in 200 μl PBS), respectively. Tumor volume was determined 3 times weekly by direct measurement with the caliper and calculated by the formula: volume V = 0.5 × a × b2 with “a” being the length and “b” the width of the tumor lump. The mice were sacrificed at the end of 4 weeks since tumor cells injection.
Statistical analysis
Values are presented as the mean ± SD (standard deviation). One-way ANOVA was carried out for multiple comparisons and two-tailed t tests were used for single comparisons. p < 0.05 was considered statistically significant. All experiments were repeated at least 3 times.
Results
Transformation of RG glioblastoma cells under long-term exposure to imatinib
While co-culturing RG glioblastoma cells, which originated from primary culture from tumor biopsy of a patient suffered from GBM [16], with 10 μM of imatinib for up to 3 months, we observed significant cell transformation and named the established cell line as RG-IM (“Materials and methods” section). Data from the cell cycle analysis indicated that, while RG cells and the same cells treated with imatinib for 48 h kept diploid and showed irregular portions of G1, S, and G2/M phases in cell cycle distribution, the RG-IM became haploid and pointed to a normal-like cell cycle distribution (Fig. 1a).
Long-term exposure to imatinib-induced RG glioblastoma cells transformation. a Cell cycle analysis in RG, RG treated with 10 μM of imatinib for 48 h, and RG-IM cells. b Comparison in morphology among RG, RG-IM, and RG treated with 1 μM of ATRA for 4 days. The pictures were taken in the same focus level with phase-contrast microscope at 100× magnifications. c The expression of GFAP and β-Tubulin III (Tuj1) detected by immunofluorescence. Immunofluorescence staining confirmed a significant upregulation of astrocyte marker GFAP and neuron marker Tuj1 in RG-IM cells compared to the RG cells. Representative images from 2 independent experiments with similar results are shown. Scale bars 100 μm
In addition, microscopic observation on RG-IM cells revealed major alteration in morphology. Unlike mainly polygonal appearance in RG cells, RG-IM culture displayed a variety of cell shapes, for example, some cells showed similar to that of mature astrocytes with stellar-shaped cell bodies and much longer, fine, and tapering processes (Fig. 1b), while certain cells exhibited enlarged cell bodies and strikingly elaborate neurite extensions with extremely long processes, often surrounded by a group of glia-like cells (Fig. 1b, c). All-trans retinoic acid (ATRA), previously used as a strong cell differentiation agent on established glioma cell lines of different origins [18] and stem like glioma cells [19], was employed as a positive control to compare its cell differentiation effect with that of imatinib on RG cells. Data indicated that, while the control cells were flattened and polygonal-shaped, the RG cells treated with 1 μM of ATRA displayed cell differentiation characteristics of a stellar shape with pronounced elongation of filamentous processes. Some of the individual cells bared similar elaborate neurite extensions with that showed in RG-IM (Fig. 1b). Further immunofluorescent microscopy revealed a significant upregulated expression of glial fibrillary acid protein (GFAP), a marker for mature astrocytes; and class III β-tubulin isotype (Tuj1), a marker for neurons in RG-IM compared to that in RG cells (Fig. 1c). These findings indicated that long-term exposure to imatinib might induce RG cells differentiation into either glia- or neuron-like cells.
PDGFR and c-Kit signaling contributed to the maintenance of undifferentiated cells in RG glioblastoma cells in vitro
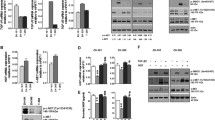
Consistent with the results from microscopic experiments (Fig. 1b, c), the data from real-time PCR indicated that long-term exposure to imatinib induced the elevated expression of neuron markers ENO2, MAP2, and astrocytic markers GFAP, S-100; however, reduced expression of stem cell-related markers, e.g., CD133, Oct-3/4, nestin, and Bmi1 in RG-IM, compared to that in parental cells (Fig. 2a; Table 2). In parallel, treatment with ATRA in RG cells caused increased expression of S100, ENO2, while decreased expression of MAP2 and GFAP (Fig. 2a). Furthermore, the expression of GFAP, S100, MAP2, and ENO2 was again upregulated in RG-IM cells treated with ATRA (Fig. 2a). These data showed potentials of both imatinib and ATRA to induce cell differentiation in RG and/or RG-IM cells.
Long-term exposure to imatinib-induced RG cells differentiation. a The relative expression of neuronal markers including ENO2, MAP2; and glial markers, S100, and GFAP at the mRNA level in RG cells treated with 1 μM of ATRA for 4 days (RG ATRA), RG-IM, and RG-IM treated with 1 μM of ATRA for 4 days (RG-IM ATRA). The relative gene expression was compared to that in RG cells. The expression of house-keeping genes GAPDH and β-actin were used to normalize variable sample loading. The data shown are the mean ± SD of 3 independent experiments. b Western blotting indicated that phosphorylation of PDGFRB and c-Kit was effectively suppressed in RG-IM cells which maintained in medium containing 10 μM imatinib for about 3 months. c Relative expression of SCF, PDGF-B, CD133, Bmi1, Oct-3/4, and S100 at the mRNA level in RG-IM cells incubated with PDGF-B (50 ng/ml) and SCF (100 ng/ml) for 4 days without serum as compared to that in RG-IM cells. Results were mean ± SD (n = 3). d The staining of nestin, the marker for neural stem cell by immunofluorescent microscopy was compared between RG and RG-IM cells. Bar 100 μm. e Significantly downregulated nestin expression at the protein level in RG-IM cells compared to that in RG cells by western blotting. Data are represented as 2–3 independent experiments. f The relative expression of PDGFRB and c-Kit at the mRNA level in RG, RG treated with 10 μM of imatinib for 4 days and RG-IM cells. Data are represented as mean values of 3 independent experiments. Bars indicate standard deviations of the mean. g Significant effect of combined gene knockdown by the application of siRNA which is specific against c-Kit and PDGFRB in RG cells. RG cells were transfected with 75 nM siRNA against PDGFRB and 25 nM siRNA against c-Kit for 48 h. Reduced PDGFRB and c-Kit expression by siRNA was shown by PCR analysis. h siRNA against PDGFRB and c-Kit significantly reduced PDGFRB and c-Kit expression at the protein level in RG cells
As a specific tyrosine kinase inhibitor, imatinib treatment inhibited the phosphorylation and thus kinase activities of PDGFR and c-Kit in glioma cells including RG cells (Fig. 2b). To further examine if cell differentiation in RG cells may result from the extended inhibition of PDGFR and/or c-kit activities by imatinib treatment (Fig. 2b), RG-IM cells were maintained with the cognate ligand of PDGFRA and PDGFRB, PDGF-B (50 ng/ml), alone or in combination with SCF (100 ng/ml), the ligand of c-Kit for 4 days in culture medium without serum, for re-stimulating the activation of PDGFR and/or c-Kit (data not shown). The real-time PCR results indicated that RG-IM cells treated with PDGF-B and SCF expressed elevated stem cell markers, such as CD133, Bmi1, Oct-3/4, and reduced astrocyte marker S100 (Fig. 2c), as compared to the same cells without treatment. Furthermore, parallel to the data from immunofluorescent staining on GFAP and Tuj1 in both RG and RG-IM cells (Fig. 1c), significantly decreased stem cell marker nestin was observed in RG-IM cells as compared to that in RG cells by immunofluorescent microscopy and Western blotting (Fig. 2d, e). However, the expression of CD133, another stem cell marker at the protein level kept comparable between RG and RG-IM cells (data not shown).
Moreover, as continuous treatment with imatinib resulted in significantly downregulated expression of the drug target kinases including PDGFRB and c-Kit at the mRNA and protein levels (Fig. 2b, f), specific siRNA on PDGFRB or c-Kit, alone or in combination in RG cells was used to observe the influences on the expression of related cell markers (Fig. 2g, h) in RG cells. The results showed that, while single siRNA application resulted in only minor alteration in the expression of cancer stem cell and terminally differentiated neural cell markers in RG cells, the double gene knockdown led to significant change on the expression of cell markers related to cancer stem cells and tumor malignancy, such as CD133, CHI3L1, SPARC, MMP1, 7, MYC, etc., and terminal neural cell markers such as GFAP and S100; although the direction of gene up- or downregulation was not exactly the same with that in RG-IM cells (Table 2). Such data indicated that combined gene knockdown of PDGFR and c-kit may significantly relate to the cancer stem cell ability in RG cells.
Decreased ability to form aggregates, colonies in vitro and tumor in vivo in RG-IM cells
Because RG-IM cells showed increased expression of terminal differentiated neural cell markers and decreased expression of markers related to cancer stem cells and tumor malignancy as compared to RG cells (Table 2), we next compared formation of cell aggregates, colonies in vitro and tumor genesis in vivo between RG and RG-IM cells. Data revealed that, while a RG cell drop containing either 4,000 or 6,000 tumor cells steadily formed respective tumor spheres for 3 days in vitro, RG-IM cell drops initiated from 3,000 to 7,000 tumor cells failed to establish tumor cell aggregates (Fig. 3a). Consistently, data from colony formation assays indicated that, while 400, 600, and 800 RG cells formed increased number of colonies within 2 weeks, the same numbers of RG-IM cells failed to form any colonies during the same period of time (Fig. 3b). Furthermore, as 1 × 107 RG cells could steadily establish tumor lumps in vivo at nude mice within 4 weeks, the same number or even increased number of RG-IM cells formed significantly smaller to nil tumors at the same time span (Fig. 3c, d). Taken together, these data indicated that RG-IM cells showed significantly decreased tumorigenicity than RG cells both in vitro and in vivo.
Long-term exposure to imatinib reduced cancer stem cell ability in RG cells. a Drops of RG cells readily formed tumor spheres after 3 days, while RG-IM cells did not form aggregates. RG-4000 indicated a drop of the cell suspension (20 μl) containing 4,000 cells, while RG-6000 indicated that of 6,000 cells. b 400, 600, and 800 RG cells steadily formed colonies in 2-week time, while RG-IM cells did not form colonies. Each experiment was carried out in duplicate. Each set of the experiments was done 3 times. c 1 × 107 RG cells steadily formed tumor lumps, while the same number of RG-IM failed to form any significant tumor in vivo (n = 7). Photographs of nude mouse were captured in each group at 4 weeks after the cells injection (upper). A photograph of the tumors is also presented (lower). d The tumor volume was measured using a caliper in nude mice injected with RG or RG-IM cells
RG cell differentiation under imatinib treatment was mediated via activation of MAPK signaling pathways
Further, we found continuously constitutive activation of ERK (MAPK) signaling in RG cells under imatinib treatment for up to 96 h, but decreased activation of AKT from 48 to 96 h of the same treatment (Fig. 4a). In comparison to RG cells, parallel level of ERK activation, but significantly decreased AKT activation, was observed in RG-IM cells (Fig. 4a). Consistently, significant activation of ERK was also observed in RG cells treated with 1 μM of cell differentiation agent ATRA for 4 and 10 days with the level of ERK activation the most significant at 4 days treatment (Fig. 4b, c). In comparison, PDGF-B treatment resulted in downregulated activation of ERK and its downstream ribosomal protein S6 (rpS6) in RG-IM cells (Fig. 4d). These results implied that ERK signaling may be actively involved in differentiation of RG cells under the treatment with either imatinib or ATRA. For example, when cell differentiation was firmly established or stabilized in RG-IM, or in RG cells treated with ATRA for longer time such as 10 days, activation of ERK could reduce or return to the baseline level (Fig. 4b, c).
Activation of MAPK signaling pathway was involved in RG cell differentiation induced by long-term exposure to imatinib. a Sustained activation of ERK (MAPK) was induced by 10 μM of imatinib in RG cells cultured with 10 % FCS for up to 96 h. Phospho-AKT was reduced from 48 h of imatinib treatment in RG cells. p-ERK and -AKT in RG-IM kept the same with that in RG cells without imatinib treatment. b, c RG cells were incubated with 1 μM cell differentiation agent, ATRA for 4 and 10 days. Protein was extracted and blotted with antibodies against total versus p-AKT (ser308), p-ERK, and p-rpS6 (ser235/6). Relative phospho-protein expression was measured as indicated in “Materials and methods” section. Data are represented as mean values of 2–3 independent experiments. Bars indicate standard deviations of the mean. Asterisks (*) indicate statistical significance (*p < 0.05, **p < 0.01). d, Expression of total versus p-ERK, p-AKT, and rpS6 in RG-IM cells treated with PDGF-B (50 ng/ml) for 4 days without serum by western blotting. e Morphological alteration in RG cells treated with ATRA, alone or in combination with protein kinase inhibitors. For single drug treatment, 1 μM of ATRA, 15 μM of MEK1/2 inhibitor U0126, or 15 μM of PI3K inhibitor LY294002, or 0.5 μM PI3K/mTOR inhibitor PI103 were used and applied for 72 h. For drug combination treatment, RG cells were pretreated with 1 μM ATRA for 24 h and then treated with U0126 (or LY294002, or PI103) for an additional 72 h before harvest for cell morphology analysis (100× magnifications) by microscopy. Western blotting was used to evaluate the relative expression of p-ERK compared to ERK in different groups. β-actin was used as a loading control. f Disturbance on forming cell aggregates in RG cells by MEK1/2 inhibitor U0126. Hanging-drop method (“Materials and methods” section) was used to observe the formation of cell aggregates in vitro in RG cells pretreated with either U0126 or LY294002. Results shown are from a representative experiment repeated at least twice with similar results
The above data were additionally confirmed by morphological observation under microscopes which showed that, while single drug treatment with U0126, an inhibitor against MEK1/2, which is the immediate upstream effectors of ERK, resulted in more flattened polygonal shape-like cells; the combination treatment with U0126 and ATRA changed the long spindle-like cells treated by ATRA alone into relatively flattened polygonal cells with shorter-processes in RG cells (Fig. 4e). On the other hand, however, we did not observe significant alteration in RG cell morphologies under the treatment of a PI3K inhibitor, LY (LY294002), or a PI3K/mTOR inhibitor, PI103, either alone or in combination with ATRA (Fig. 4e). Moreover, we demonstrated that RG cell drops which were pretreated with different concentrations of U0126, but not that of LY, for 24 h failed to form tumor cell aggregates in vitro (Fig. 4f).
Discussion
In this study, we demonstrated induction of cell differentiation as well as reduced cancer stem cell ability in RG glioblastoma cells with long-term exposure to imatinib, a tyrosine kinase inhibitor against PDGFR and c-Kit. The overall cell differentiation effect induced by imatinib treatment was comparable to that by cell differentiation agent ATRA in RG cells. Furthermore, we found that the cell differentiation was mainly driven by MAPK (ERK) signaling pathways downstream of PDGFR and c-Kit. Our findings provided evidences that PDGFR and c-Kit signaling may play an important role in the maintenance of the glioma stem cells in vitro, which may represent a subpopulation of the GBM cells in culture; targeted inhibition on such cellular signaling by a kinase inhibitor such as imatinib might thus have the potential to reduce malignancy of human glioblastoma.
Research data indicated that GBM cells could differentiate not only into neuronal and glial cells but also into non-neural cells [20], revealing the plasticity of GBM cells under specific environmental stimuli. Accumulated evidence suggested that PDGF/R signaling may play an important role in tumorigenesis of the brain tumor [3]. Erlandsson et al. [5] recently found that when PDGFR signaling was blocked during differentiation of neural stem/progenitor cells, neurons, and oligodendrocytes differentiated more rapidly. These and our data pointed out that PDGFR signaling may be vital to the maintenance of stem cell ability in brain tumor. On the other hand, although SCF/c-Kit signaling was found to mainly involve in hematopoiesis, it was recently demonstrated that such signaling promoted the migration and proliferation of mesenchymal and neural stem cells in vitro [21]. In our results, while the cell differentiation induced by long-term exposure to imatinib was not found in GBM cell lines other than RG, these included T98G and U87MG cells which expressed only PDGF/R but not c-Kit (data not shown); application of double siRNA against PDGFRB and c-Kit in RG cells resulted in significant alteration on the expression of cancer stem cell-related genes. Therefore, our data further indicated that co-operation between PDGFR and c-Kit signaling played an essential role in cancer stem cell ability in RG cells.
While dedifferentiated mature astrocytes were indicated as one of the sources of GSC in GBM [2], genetic alterations other than PDGFR/c-Kit were also linked with undifferentiated state of astrocytes in high grade glioma. For example, the mesenchymal subtype, one of the molecular subclasses of GBM, was characterized by high expression of CHI3L1 (Chitinase 3-Like-1, also known as YKL40) [22], which was a typical mesenchymal marker and the over-expression of which was also particularly related to recurrent glioblastomas. The MYC proto-oncogene was over-expressed and associated with an undifferentiated state in GBM [23]. It was shown that MYC enhanced the sensitivity of GFAP-expressing astrocytes to gliomagenesis [24]. Furthermore, the expression of stem cell marker, Oct-3/4 (also known as POU5F1) was shown in various human solid tumors and cancer cell lines, and higher expression correlated with tumor progression, malignancy, and prognosis [25]. Du et al. [26] recently reported that the expression levels of Oct-3/4 correlated with the grade of gliomas, and that Oct-3/4 was necessary to maintain rat C6 glioma cells in an undifferentiated state. Moreover, it was indicated that Oct-3/4 upregulated matrix metalloproteinase-2 (MMP-2), MMP-9, and MMP-13 production, which could contribute to tumor metastasis and invasion in bladder cancer [27]. In our results, we found reduced expression of mesenchymal markers (CHI3L1, FN, SPARC), MYC, Oct-3/4, and CD133, a typical cancer stem cell marker, in addition to MMP1, MMP7 at the mRNA level in RG-IM cells. Furthermore, RG-IM cells were unable to form tumor spheres in culture and bore significantly reduced tumorigenicity as compared to parental RG cells in vivo. We thus speculate that long-term exposure to imatinib might decrease malignancy in RG glioblastoma cells.
Mitogens such as EGF, FGF, PDGF and thereof related signaling pathways promote the growth of adult neural progenitor cells [2]. Constitutive activation of growth factor receptor signaling and their downstream signaling pathways, including canonical Ras/Raf/MAPK and PI3K/AKT signaling is a hallmark of malignant glioma. Therefore, it is highly likely that GSC in GBM cells co-opt with mitogenic cues that regulate the growth of normal neural progenitors. Thus far, there were only a few studies analyzing changes in cellular signaling pathways on glioma cell differentiation [28]. Recent data indicated that both the activity of the PI3K/AKT pathway and expression of stem cell markers correlated with aggressive behavior and resistance to therapy in human high grade glioma [22, 29]. In mouse models of GBM, nestin-expressing stem cells have a high level of AKT pathway activity [30]. However, our data demonstrated that cell differentiation in RG glioblastoma cells treated by imatinib was mainly driven by MAPK/ERK signaling pathway rather than PI3K/AKT pathways, and the same effect was confirmed in the glioblastoma cells treated with the cell differentiation agent ATRA. Self-renewal pathways such as the NOTCH and BMP signaling cascades could be of interest because they are involved in the maintenance of stem cell properties. Yet, we did not observe alteration on the expression of NOTCH or BMP pathways in RG cells with long-term exposure to imatinib.
Imatinib is well tolerated and highly effective in treating chronic myeloid leukemia (CML) by targeting the constitutively active BCR-ABL kinase. Recently, it was indicated that long-term imatinib therapy for up to 3 years promoted bone formation by induction of an increase in osteoblast differentiation and function [31], and reduced the abundance of leukemic stem cells [32] in CML patients. Previous reports showed that imatinib treatment inhibited neurosphere formation and tumor growth of glioblastoma stem cells [33]. Nevertheless, GBM is highly complex and heterogeneous in nature [2]. This property mostly accounts for the modest anti-tumor effect in patients with GBM treated by targeted agents including imatinib [14]. We observed differential effect in different PDGF/R-expressing glioblastoma cells under the treatment of imatinib, including U87MG, T98G, and RG cells (data not shown). Moreover, cancer stem cells might contribute to resistance to radiotherapy and chemotherapy in bulk of cancer; it was indicated that imatinib sensitized malignant glioma cells to radiotherapy [34], and combined therapy with other chemotherapeutic agents, e.g., Hydroxyurea [35]. Therefore, our results might present possible mechanisms involved in the enhanced anti-tumor effect by a combination therapy with imatinib in patients with malignant glioma. However, further molecular characterization of RG cells may define subtypes of GBM that are suitable for imatinib treatment; discovery of such biological properties of GBM might provide considerable information on the development of new therapeutic strategies aimed at forcing glioblastoma stem cell differentiation. In conclusion, targeted inhibition on PDGFR and c-Kit signaling disturbed glioma stem cell biology in subsets of GBM cells and may have potentials in clinical applications.
References
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760
Stiles CD, Rowitch DH (2008) Glioma stem cells: a midterm exam. Neuron 58:832–846
Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, Nister M (1992) Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res 52:3213–3219
Varela M, Ranuncolo SM, Morand A, Lastiri J, De Kier Joffe EB, Puricelli LI, Pallotta MG (2004) EGF-R and PDGF-R, but not bcl-2, overexpression predict overall survival in patients with low-grade astrocytomas. J Surg Oncol 86:34–40
Erlandsson A, Brannvall K, Gustafsdottir S, Westermark B, Forsberg-Nilsson K (2006) Autocrine/paracrine platelet-derived growth factor regulates proliferation of neural progenitor cells. Cancer Res 66:8042–8048
Uhrbom L, Hesselager G, Nister M, Westermark B (1998) Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res 58:5275–5279
Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC (2001) PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev 15:1913–1925
Blom T, Fox H, Angers-Loustau A, Peltonen K, Kerosuo L, Wartiovaara K, Linja M, Janne OA, Kovanen P, Haapasalo H, Nupponen NN (2008) KIT overexpression induces proliferation in astrocytes in an imatinib-responsive manner and associates with proliferation index in gliomas. Int J Cancer 123:793–800
Sihto H, Sarlomo-Rikala M, Tynninen O, Tanner M, Andersson LC, Franssila K, Nupponen NN, Joensuu H (2005) KIT and platelet-derived growth factor receptor alpha tyrosine kinase gene mutations and KIT amplifications in human solid tumors. J Clin Oncol 23:49–57
George D (2001) Platelet-derived growth factor receptors: a therapeutic target in solid tumors. Semin Oncol 28:27–33
Buchdunger E, Zimmermann J, Mett H, Meyer T, Muller M, Druker BJ, Lydon NB (1996) Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res 56:100–104
Demetri GD, von MM, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H (2002) Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 347:472–480
Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NC, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M, Wlodarska I, Kantarjian H, Marynen P, Coutre SE, Stone R, Gilliland DG (2003) A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med 348:1201–1214
Wen PY, Yung WK, Lamborn KR, Dahia PL, Wang Y, Peng B, Abrey LE, Raizer J, Cloughesy TF, Fink K, Gilbert M, Chang S, Junck L, Schiff D, Lieberman F, Fine HA, Mehta M, Robins HI, DeAngelis LM, Groves MD, Puduvalli VK, Levin V, Conrad C, Maher EA, Aldape K, Hayes M, Letvak L, Egorin MJ, Capdeville R, Kaplan R, Murgo AJ, Stiles C, Prados MD (2006) Phase I/II study of imatinib mesylate for recurrent malignant gliomas: north American Brain Tumor Consortium Study 99–08. Clin Cancer Res 12:4899–4907
Dong Y, Jia L, Wang X, Tan X, Xu J, Deng Z, Jiang T, Rainov NG, Li B, Ren H (2011) Selective inhibition of PDGFR by imatinib elicits the sustained activation of ERK and downstream receptor signaling in malignant glioma cells. Int J Oncol 38:555–569
Ren H, Tan X, Dong Y, Giese A, Chou TC, Rainov N, Yang B (2009) Differential effect of imatinib and synergism of combination treatment with chemotherapeutic agents in malignant glioma cells. Basic Clin Pharmacol Toxicol 104:241–252
Rungarunlert S, Techakumphu M, Pirity MK, Dinnyes A (2009) Embryoid body formation from embryonic and induced pluripotent stem cells: benefits of bioreactors. World J Stem Cells 1:11–21
Das A, Banik NL, Ray SK (2008) Retinoids induced astrocytic differentiation with down regulation of telomerase activity and enhanced sensitivity to taxol for apoptosis in human glioblastoma T98G and U87MG cells. J Neurooncol 87:9–22
Campos B, Wan F, Farhadi M, Ernst A, Zeppernick F, Tagscherer KE, Ahmadi R, Lohr J, Dictus C, Gdynia G, Combs SE, Goidts V, Helmke BM, Eckstein V, Roth W, Beckhove P, Lichter P, Unterberg A, Radlwimmer B, Herold-Mende C (2010) Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer Res 16:2715–2728
Ricci-Vitiani L, Pallini R, Larocca LM, Lombardi DG, Signore M, Pierconti F, Petrucci G, Montano N, Maira G, De MR (2008) Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ 15:1491–1498
Bantubungi K, Blum D, Cuvelier L, Wislet-Gendebien S, Rogister B, Brouillet E, Schiffmann SN (2008) Stem cell factor and mesenchymal and neural stem cell transplantation in a rat model of Huntington’s disease. Mol Cell Neurosci 37:454–470
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173
Hoshimaru M, Ray J, Sah DW, Gage FH (1996) Differentiation of the immortalized adult neuronal progenitor cell line HC2S2 into neurons by regulatable suppression of the v-myc oncogene. Proc Natl Acad Sci USA 93:1518–1523
Lassman AB, Dai C, Fuller GN, Vickers AJ, Holland EC (2004) Overexpression of c-MYC promotes an undifferentiated phenotype in cultured astrocytes and allows elevated Ras and Akt signaling to induce gliomas from GFAP-expressing cells in mice. Neuron Glia Biol 1:157–163
Monk M, Holding C (2001) Human embryonic genes re-expressed in cancer cells. Oncogene 20:8085–8091
Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, Cao X, Ling EA, Hao A (2009) Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia 57:724–733
Chang CC, Shieh GS, Wu P, Lin CC, Shiau AL, Wu CL (2008) Oct-3/4 expression reflects tumor progression and regulates motility of bladder cancer cells. Cancer Res 68:6281–6291
Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL (2006) Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 444:761–765
Beier D, Wischhusen J, Dietmaier W, Hau P, Proescholdt M, Brawanski A, Bogdahn U, Beier CP (2008) CD133 expression and cancer stem cells predict prognosis in high-grade oligodendroglial tumors. Brain Pathol 18:370–377
Hambardzumyan D, Squatrito M, Carbajal E, Holland EC (2008) Glioma formation, cancer stem cells, and akt signaling. Stem Cell Rev 4:203–210
Fitter S, Dewar AL, Kostakis P, To LB, Hughes TP, Roberts MM, Lynch K, Vernon-Roberts B, Zannettino AC (2008) Long-term imatinib therapy promotes bone formation in CML patients. Blood 111:2538–2547
Tang M, Gonen M, Quintas-Cardama A, Cortes J, Kantarjian H, Field C, Hughes TP, Branford S, Michor F (2011) Dynamics of chronic myeloid leukemia response to long-term targeted therapy reveal treatment effects on leukemic stem cells. Blood 118:1622–1631
Gal H, Pandi G, Kanner AA, Ram Z, Lithwick-Yanai G, Amariglio N, Rechavi G, Givol D (2008) MIR-451 and imatinib mesylate inhibit tumor growth of glioblastoma stem cells. Biochem Biophys Res Commun 376:86–90
Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, Tofilon PJ (2003) Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res 63:7377–7383
Reardon DA, Egorin MJ, Quinn JA, Rich JN, Gururangan S, Vredenburgh JJ, Desjardins A, Sathornsumetee S, Provenzale JM, Herndon JE, Dowell JM, Badruddoja MA, McLendon RE, Lagattuta TF, Kicielinski KP, Dresemann G, Sampson JH, Friedman AH, Salvado AJ, Friedman HS (2005) Phase II study of imatinib mesylate plus hydroxyurea in adults with recurrent glioblastoma multiforme. J Clin Oncol 23:9359–9368
Acknowledgments
The presented research project was supported by the Key Project Science Foundation of Heilongjiang Province, China (Grant-ZD200804-01), Chinese Postdoctoral Fellowship (Grant-2008043938), and National Science Foundation of China (Grant NFSC-30227738), to Huan Ren.
Conflict of interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dong, Y., Han, Q., Zou, Y. et al. Long-term exposure to imatinib reduced cancer stem cell ability through induction of cell differentiation via activation of MAPK signaling in glioblastoma cells. Mol Cell Biochem 370, 89–102 (2012). https://doi.org/10.1007/s11010-012-1401-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-012-1401-0