Abstract
Linking spatial pattern and process is a difficult task in landscape ecology because spatial patterns of populations result from complex factors such as individual traits, the spatio-temporal variation of the habitat, and the relationships between the target species and other species. Mechanistic models provide tools to bridge this gap but they are seldom used to study the influence of landscape patterns on biological processes. In this paper, we develop a methodological approach based on sensitivity and multivariate analyses to investigate the relationship between the biological parameters of species and landscape characteristics. As a case study, we used a tritrophic system that includes a host plant (oilseed rape, Brassica napus L.), a pest of the host plant (the pollen beetle, Meligethes aeneus F.), and the main parasitoid of the pest (Tersilochus heterocerus). This tritrophic system was recently represented by a model (Mosaic-Pest) that is spatially explicit at the landscape scale and that includes 32 biological parameters. In the current study, model simulations were compared with observed data from 35 landscapes differing in configuration. Sensitivity analysis using the Morris method identified those biological parameters that were highly sensitive to landscape configuration. Then, multivariate analyses revealed how a parameter’s influence on model output could be affected by landscape composition. Comparison of simulated and observed data helped us decrease the uncertainty surrounding the estimated values of the literature-derived parameters describing beetle dispersal and stage transition of the parasitoid at emergence. The advantages of using multivariate sensitivity analyses to disentangle the links between patterns and processes in landscape-scale spatially explicit models are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spatially-explicit simulation models are powerful tools for understanding the links between patterns and processes (Grimm et al. 2005; McIntire and Fajardo 2009). Because spatial models explicitly consider the processes responsible for the observed patterns, i.e. spatio-temporal variation of the population of a given species itself (Tscharntke and Brandl 2004), or in interaction with other species (Bianchi and Schellhorn 2009) they can be used as virtual laboratories (Charnell 2008) to test the effects of a given set of processes on a spatial pattern. Thus, the effects of habitat heterogeneity (Frair et al. 2005), demographic parameters and dispersal (Wiegand et al. 2004), or gene flow (Viaud et al. 2008) on populations patterns have been successfully studied using spatially-explicit simulation models. However, simulation models require often many parameters to represent the complexity of the simulated system that is necessary to respond to the biological question.
To better predict the real trends of the system, spatially-explicit simulation models need to be parameterized on the basis of one of several observed dataset. Optimizing the match between observed and simulated system responses is done via inverse modelling (Kramer-Schadt et al. 2007), i.e. searching the model parameterizations that fit the observed data best. Optimization tools suitable for use with spatially-explicit simulation models are scarce because model complexity generally makes conventional statistical techniques (e.g., sum of squares, maximum likelihood) unusable for parameter estimation. Bayesian tools such as approximate Bayesian computation are alternatives but require a large number of simulations to be efficient, which makes the study of spatially-explicit simulation models computationally expensive (Csilléry et al. 2010; Hartig et al. 2011). Inverse modelling techniques have been used with success to calibrate a large range of spatially-explicit simulation models, generally in association with a global sensitivity analysis (Kramer-Schadt et al. 2004; Wiegand et al. 2004; Beaudouin et al. 2008; Martinez et al. 2011).
The modelling of the processes underlying spatial patterns is greatly affected by parameter estimation because of non-linear dependencies, threshold effects, and/or negative feedbacks. Consequently, it is crucial to use accurate parameter estimates in order to point the information which is most lacking for further reducing uncertainty in model predictions (Wiegand et al. 2004). Because not all parameters have the same influence on model output, it can be useful to determine whether uncertainties in particular parameter values lead to large uncertainties in the output, i.e., it is useful to determine which parameter estimates warrant the most attention. This is done with sensitivity analysis, which measures the impact of input factors on a selected output (Saltelli et al. 2000). Several methods are available for sensitivity analysis and can be classified into two categories: local sensitivity analyses (one-at-a-time methods, such as FAST or Sobol’ methods), in which the effect of the variation of a single factor is estimated, and global sensitivity analyses (e.g., the Morris method), in which the output of a factor is studied when all the factors are varying (Cariboni et al. 2007). Selection of sensitivity analysis method depends first on the scientific question asked, then on the number of parameters under consideration and on the existence of non-linear effects or interactions between parameter effects. The use of sensitivity analysis to investigate how uncertainties in parameter values affect a model’s inferential power is an integral part of modelling (Cariboni et al. 2007).
However, an additional difficulty in studying spatially-explicit simulation models is that the biological processes may respond differently depending on landscape composition and structure. For example, the importance of dispersal may be greater in fragmented than in homogeneous landscapes (Vinatier et al. 2012b). Disentangling the link between ecological processes and landscape composition and structure has been of particular importance in landscape ecology, for example, respect to the question of the relative impact of habitat loss and fragmentation on population dynamics (Wiegand et al. 1999, 2005; Fahrig 2003). As before, sensitivity analyses can provide insight because such analyses provide a convenient framework for understanding the behaviour of complex mechanistic models. However, sensitivity analysis of spatially-explicit simulation models is especially challenging because there is no straightforward method to include landscape characteristics into the analysis.
We present here a new approach for disentangling the complex link between ecological processes and landscape composition and structure. The approach combines sensitivity and multivariate analyses of a spatially-explicit simulation model. To demonstrate this approach, we use a tritrophic system that includes a pest, its parasitoid, and its host plant and for which a spatio-temporal model has been developed (Vinatier et al. 2012a). The development of the model, called Mosaic-Pest, was motivated by the need to design landscape-scale methods to control an important pest of oilseed rape, the pollen beetle Meligethes aeneus, and also to manage an important parasitoid of the beetle, Tersilochus heterocerus. Mosaic-Pest considers landscape composition because both the pollen beetle and parasitoid are sensitive to the proportion of semi-natural habitats (i.e., woodlands and grasslands), which are overwintering sites for the pollen beetle and nectar sources for the parasitoid (Rusch et al. 2011). Mosaic-Pest also considers agricultural practices that affect pest densities, such as crop rotation (Rusch et al. 2011), ploughing (Nilsson 2010) and precocity of rape varieties (Cook et al. 2007). The choice of this case study was also motivated by the availability of previously collected data on pollen beetle densities and parasitism rates in farmer fields located at the centre of 35 maps differing in landscape composition and structure (Rusch et al. 2011).
The overall goals of this research were to develop a general method for studying the relationship between parameters and landscape composition and structure with spatially-explicit simulation models and to develop a general method for improving the estimation of parameter values. With respect to our tritrophic study system, the specific objectives were (i) to assess via a global sensitivity analysis the impact of parameter uncertainties on population densities of pollen beetles and parasitism rates, (ii) to narrow the range of confidence intervals for parameters obtained in the literature by using a variance decomposition technique for the comparison of simulated and observed data, and (iii) to determine how the main effects of biological parameters vary with landscape composition via sensitivity and multivariate analyses.
Methods
Overview of the model
The Mosaic-Pest model was described in detail by Vinatier et al. (2012a) and in Appendix 1 (Supplementary Material). It simultaneously represents crop planting and development, and host and parasitoid dynamics and their interactions. The model is spatially explicit, i.e., it is based on a 100 × 100 grid of 50-m cells. The model considers four different habitat types (oilseed rape fields, previous oilseed rape fields, woodlands, and grasslands) because of their different influences on insect overwintering, feeding, and egg laying. The model was developed with Netlogo software (Wilensky 1999).
In the Mosaic-Pest model, populations of pollen beetles are divided into five stages: egg, 1st instar larva, 2nd instar larva, pupa, and adult. The adult changes status during its life cycle from dispersing towards overwintering sites, overwintering, dispersing for feeding, dispersing for egg laying, to egg laying and finally to death. Populations of parasitoids are divided into immature and adult stages, and adult parasitoids change status as described for pollen beetles, except for dispersing towards overwintering sites status. At the end of oilseed rape-growing season, the new generation of parasitoids remains as diapausing adults within host cocoons in the soil, whereas new adults of pollen beetles disperse to the overwintering sites, i.e., woodlands. The size of populations of each species varies with time according to transition probabilities depending on their stage, status, and location. Parasitism of pollen beetles is described by a Thompson model (Jourdheuil 1960; Mills and Getz 1996) that represents the functional response of the parasitoid, i.e. the rate at which the hosts are parasitized as a function of hosts and parasitoids densities. Dispersion events of populations occur according to a cell-to-cell redistribution mechanism, depending on both a dispersal kernel and relative attractiveness of habitat elements. Survival probability of adults depends on the power of the distance covered by the population during dispersal.
This model includes 32 parameters describing the demographics and dispersal of the pest and the parasitoid (Table 1). Parameters controlling demographic processes are densities of insects in overwintering sites (N Ma0 ), temperature threshold for emergence (θMa), proportion of the population surviving at emergence (πTh,immature,NT or πTh,immature,ploughing if ploughing is applied) and at each stage transition (πMa,adult,egg,larva,or pupa), and duration of each stage (δ Ma,adult,egg,larva, or pupa). Parameters controlling dispersal are the maximum dispersal distance (in meters) travelled by insects before feeding (ωMa,adult), the proportion of individuals that survive per meter travelled (τMa), and the decrease in habitat attractiveness with distance travelled (βMa,adult). Values for most of the parameters were obtained from the literature (Table 1), but some parameters were not present in the literature and others were present but with a large uncertainty around the estimated values.
Observed data used for model initialisation and optimization
The realism of the Mosaic-Pest model was increased by comparing simulation data with observed data. The observed data set consisted of pollen beetle densities and parasitism rates sampled along a transect in the central field of 35 different landscapes at the end of the flowering period during the years 2008 and 2009 [for more details on sampling and observations, see Appendix 2 (Supplementary Material)].
The observed data were collected in an agricultural territory located in northwestern France (49°25′N, 1°12′E) and consisting of landscapes of arable land, small woodland fragments, hedgerows, and grasslands. We considered 35 non-overlapping maps of 2.5-km radius centred on an oilseed rape field within the region. The maps were generated from aerial photographs (BD ORTHO®, IGN, 2004). Each map contained different proportions of winter oilseed rape, other crops, grassland, and woodland, with landscapes ranging from simple (i.e., <5 % semi-natural habitats) to more complex (i.e., up to 58 % semi-natural habitats). Semi-natural habitats were grasslands, woodlands. Agricultural practices (i.e., crop allocation, rotation sequences, and ploughing) were determined based on intensive field inspection (Rusch et al. 2011) and on the official GIS-based system used by farmers to declare crops and apply for subsidies (Registre Parcellaire Graphique, Reglement communautaire no. 1593/2000). The landscapes were then rasterized using a 50 × 50 m resolution. Examples of the resulting maps are presented in Appendix 3 (Supplementary Material).
The simulations were conducted so that the modelled system was as similar as possible to the real conditions: GIS polygons representing cultivated fields, forests, and pastures were rasterized, and observed crop allocation, rotation sequences, and ploughing were applied to the model. For initialising insect populations, we assigned to every cell of the grid the same number of parasitoids or pollen beetles, which were at the immature stage for parasitoids and at the adult stage for pollen beetles (the initial densities were considered as a parameter in the sensitivity analysis). Every simulation began on 1 January, and simulations were run for 300 days, from the beginning of the overwintering period to the emergence of new pollen beetle adults.
Sensitivity analyses and inverse parameter estimation
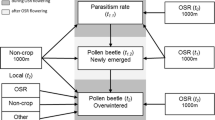
We propose a five-stage approach for improving the estimation of parameter values, analysing the sensitivity of complex models to biological parameters, and assessing interactions between influence of biological parameters and landscape conditions while keeping the number of simulations reasonably small (Fig. 1). In stage 1, well-established methods for sensitivity analysis of complex models (i.e., the Morris method) are used. In stage 2, results of the sensitivity analysis are analysed to reduce the range of uncertainty surrounding important and poorly estimated biological parameters and thus to make the model more precise, accurate, and unbiased. In stage 3, the most influential parameters are selected on the basis of the Morris method. In stage 4, parameters that are similarly influenced by the landscape are identified. In step 5, landscape characteristics that substantially affect the key parameters’ influences are determined. Statistical analyses were conducted with R software (R Development Core Team 2010) using the base, the package “sensitivity” for the construction of the sensitivity plan, the package “Ade4” for multivariate analyses, and the package “RNetlogo” to embed Netlogo into the R environment (Thiele and Grimm 2010).
Overview of the five-stage approach used in the study. Pi, Li, μi, and Mi correspond to the abbreviate of the ith parameter, landscape, Morris influence, and landscape metric, respectively. Black boxes illustrate the objective (in italics) of each intermediate stage of the approach. Thin arrows illustrate the transition steps of the approach. Large coloured arrows illustrate (i) the statistical (ANOVA, PCA and co-inertia for Principal Component and co-inertia analyses, respectively, correlation for multiple correlation using Pearson’s coefficient)and (ii) the mechanistic (the Mosaic-Pest logo figures the model functioning) parts of the approach. (Color figure online)
The Morris method for sensitivity analysis of biological parameters (stage 1)
For stage 1, we used the Morris screening method (Morris 1991) to identify the parameters that had the most influence on the variability of the two output variables in the observed data set, which were densities of pollen beetles and parasitism rates. The Morris method is suitable for models that have many input parameters and that are computationally expensive (Cariboni et al. 2007). We preferred the Morris method over other parameter search methods, such as the Latin hypercube sampling, because the latter encounters computational limitations when number of parameters exceeded 10–15 (Hartig et al. 2011). The Morris method identifies influential parameters with a relatively low number of model evaluations.
Following the Morris method, the possible range (defined according to the uncertainty surrounding estimates in the literature) of each of the 32 parameters was divided into five levels with a resolution ∆ (see Table 1). A starting point was defined by sampling a set of start values within the possible values for all parameters. From this point, a trajectory was defined by increasing (or decreasing) each parameter in turn with a step size of ∆ and the model was run at each of the k + 1 steps of the trajectory. This procedure was repeated 40 times to explore a significant fraction of the total uncertainty space, thus leading to 40 × (32 + 1) = 1,320 sensitivity runs. Furthermore, each sensitivity run is repeated for the 35 maps, leading to 1,320 × 35 = 46,200 simulations.
The elementary effect (EE i ) of a parameter θ k on a trajectory j was calculated as:
with e i = ±1 and y j the model output.
The mean (μ*) and the standard deviation (σ) of the absolute values of the elementary effects over the trajectories were used as sensitivity measures to ascertain the importance of the factors. A large μ* indicates a large overall influence of the parameter and a large σ implies a dependency of the parameter on the value of the other parameters through non-linear or interaction effects.
Comparison between observed and simulated data (inverse modelling, stage 2)
For stage 2, we selected among the 1,320 tested parameterizations those that optimized the relationship between observed data and simulated data for the two output variables (pollen beetle densities and parasitism rates) across all landscape maps. This was necessary because no single parameterization yielded a good fit of the model to both output variables. We preferred considering this relationship rather than comparing directly the squared differences between predicted and observed variables because the model is devoted to exploratory analyses, i.e. our objective was to analyse how the system responds to landscape configuration, rather than prediction analyses, i.e. predicting without error the response variables for a given map. Consequently, bias was not considered a problem and precision was less important than accuracy. For each sensitivity run, which corresponds to a set of 35 simulated map values, we fitted a linear model on simulated versus observed data for pollen beetle and parasitism rate separately. The linear model of the form Y = a X + b was used to estimate the realism of the Mosaic-Pest model, where Y = simulation data, X = observation data. Good accuracy leads to a slope close to 1, good precision leads to a high R2 and absence of bias is reflected, when the slope is close to one, by the intercept being close to 0.
We first selected the parameterization for which (i) the linear model was significant (P < 0.05) for both output variables in the data set, (ii) the slope and the intercept were non-significantly different from 1 and 0, respectively, and (iii) the value of the R2 was higher than 0.2. On the basis of this first selection of parameters’ combinations, we selected the unique combination of parameters that was both in the tenth highest levels of R2 for pollen beetles and parasitism rates, respectively. We preferred the linear model to the Spearman’s or the root-mean-square deviation statistics because the latter two were less discriminatory for our data set (data not shown).
We conducted a multiple ANOVA analysis on the 1,320 sensitivity runs to test the overall influence of each parameter on this linear model, taking separately the R2, intercept, and slope of the linear model as a response variable. Each biological parameter was treated as an input factor with five modalities (the five levels along the range of possible parameter values). The normality of the response variable and residuals was tested using the Shapiro–Wilk test. Following Monod et al. (2006), a sensitivity index of each factorial term (biological parameter) was calculated on the basis of its sum of squares to assess the relative influence of each parameter on the relationship between the observed and simulated data.
Selection of the most influential parameters (stage 3)
In stage 3 we identified the parameters with the strongest influence on the model outputs. To this end we determined the parameters for which the values of the Morris μ* were among the three highest for at least one map. The ordered sum of the Morris μ* for the different maps showed that for almost all maps only the three highest μ* were the most important, whereas the others were closed to zero. The Morris method enabled us to estimate the mean μ* and the standard deviation δ of the elementary effect of each parameter on the model’s outputs, i.e., densities of pollen beetles and parasitism rates.
Note that the parameters identified as influential in stages 2 and 3 were not necessarily the same because stage 3 focused on the parameters that influenced value of model output for the highest number of landscapes, whereas stage 2 focused on the parameters that influenced model accuracy and precision when compared against real observations. Because observations were made in landscapes chosen specifically to vary in terms of landscape factors (proportion of different land uses), a large effect of a biological parameter on the agreement between observations and simulations indicated that this parameter plays a key role in the relationship between landscape factors, pests, and parasitoids.
Effect of landscape composition on the influence of biological parameters (stage 4)
For stage 4, we compared the medians of the values for the Morris μ* for biological parameters in each pair of maps to determine whether the influence of parameters shared the same behaviour across landscape maps. This analysis was based on (i) multiple correlation analysis between parameter influences for a low number of key parameters, or (ii) principal component analysis (PCA) of parameters influences for a high number of key parameters (we highlighted groups of biological parameters depending on the species and the type of parameter, i.e., dispersal or demographic). The box-cox transformation (lambda = 2) was applied to the values of Morris’ μ* for normalization, as no a priori exists on the distribution of this variable.
Determining landscape characteristics that influence values of Morris μ* parameters (stage 5)
For stage 5, we used multiple correlation or multivariate analyses to understand the relationships between the values of Morris’ μ* for parameters and landscape metrics on the two outputs of the model (densities of pollen beetles and parasitism rates). The choice of multiple correlation methods or multivariate analyses depends on the number of influent parameters. In our case, we considered arbitrarily a threshold of ten parameters. For a parameters’ number exceeding this threshold, combination of correlations between variables was too important to be considered in a synthetic table whereas they have been calculated (data not shown) and the use of PCA as a more aggregative techniques was preferred.
Landscape metrics were computed on each of the 35 maps via buffer analyses (Ricci et al. 2009; Rusch et al. 2011) [Appendix 3 (Supplementary Material)]. We considered the proportion of four habitat classes (i.e., woodland, grassland, previous-year oilseed rape fields, and oilseed rape fields) in eight circular zones with radii ranging from 250 to 1750 m around the centre of each map. The matrix of landscape metrics by map (i.e., a data set of 32 Landscape metrics × 35 maps) was named the Landscape metrics data set.
For a number of key parameters lower than ten, the correlation between the value of Morris’ μ* for a given parameter and a given Landscape metric was calculated across landscapes to determine whether the influence of a given parameter on the model output depended on the conditions (in terms of landscape composition) under which it was evaluated and which habitats influenced the model’s response to biological parameters and at what scale. For a number of key parameters higher than 10, we used multivariate analyses to describe the relationship between parameters and landscape. The values of Morris’ μ* and Landscape metrics data sets were first re-projected separately using a principal component analysis (PCA) to define aggregated variables that represented groups of biological parameters and descriptors of landscape complexity. Subsequently, a co-inertia analysis was applied on the two PCAs. The co-inertia analysis is a multivariate method that identifies trends or co-relationships in multiple data sets that contain the same samples. The significance of co-inertia analyses was tested using the Monte-Carlo random permutation test. The ways in which groups of parameters (demographic or dispersal parameters for each species separately) and landscape composition descriptors (buffer radii and habitat types) were related to co-inertia axes were described.
Results
Sensitivity analysis using Morris method and selection of the most influential parameters (stage 1 and stage 3)
Of the 32 parameters of the Mosaic-Pest model that were tested, six and 19 parameters were identified as influential (i.e., they had Morris μ* values that were among the three highest for at least one map) for pollen beetle densities and parasitism rates, respectively (Table 1). The six parameters for pollen beetle density were the pest dispersal parameters ωMa,adult, τMa, and βMa and the pest demographic parameters πMa,winter, δ Ma,adult, and N Ma0 . The 19 parameters for parasitism rates included both pest and parasitoid dispersal and demographic parameters. Some parameters did not influence pollen beetle density or parasitism rate because their uncertainty level (estimated from the literature) was small and/or their influence in the model (as characterised by the Morris method) was negligible (e.g., δ Ma and πMa of pollen beetles eggs, larvae, and pupae).
Figure 2 shows the relative importance of the values of Morris’ μ* and δ for each parameter. The large size of box plots indicates that Morris μ* was highly variable across map patterns, highlighting that landscape composition of maps greatly affected parameter influence (Fig. 2a). Spread of the δ values across landscape maps was low for πMa,winter and N Ma0 parameters and high for dispersal parameters and δ Ma,adult (Fig. 2b), indicating that the interactions between other parameters and πMa,winter or N Ma0 was low regardless of the landscape composition while the importance of the interaction between the influence of dispersal parameters of δ Ma,adult and the other parameters depended on the landscape context. The boxplot medians of δ parameters among biological parameters mirrored the temporal sequence of events in the model, i.e., δ values progressively increased for parameters controlling events during the overwintering period, dispersal events, and then adult death. Because the Morris δ indicates non-linear and interaction effects, this is not surprising: the value of δ increased with the cumulative number of events.
Box plots (n = 35) for the values of the Morris a μ* and b δ across landscape maps for pollen beetle densities. Only the six most influential parameters are presented (a most influential parameter was defined as one whose value for the Morris μ* was among the three highest for at least one map). Each boxplot contains the lower whisker, the lower hinge (first quartile), the median, the upper hinge (third quartile) and the extreme of the upper whisker. The whiskers extend to the most extreme data point that is no more than 1.5 times the interquartile range from the box
Inverse modelling (stage 2)
When a linear model was fitted to the relationship between observed data and each run of the sensitivity analysis plan, R2 values ranged from 0.15 to 0.43 for pollen beetle densities and from 0.01 to 0.25 for parasitism rates. The ANOVA models accounted for more than 80 % of the variability of each response variable. Three parameters for pollen beetle density and one parameter for parasitism rate had sensitivity indices >10 % (Monod et al. 2006) (Table 1). Figure 3a shows how parameter levels of τMa and ωMa,adult affected R2, intercept, and slope values of the linear regression model, with an optimum of 0.999 m−1 for τMa and 300 m for ωMa,adult. The optimum for the parameter πTh,immature,ploughing was 0.2, i.e., the presence of ploughing destroyed 80 % of the pupae (Fig. 3b). The parameter πMa,winter was not presented in Fig. 3a because its influence on the relationship between observation and simulation was not clearly emphasized by the graph. Some parameters, such as N Ma0 , δ Ma,adult, and βMa, greatly affected pollen beetle densities as determined by the Morris method (Fig. 2) but had little effect on the agreement between the simulated and observed pollen beetle data sets [sensitivity indices below 5 % (Monod et al. 2006)], indicating that these parameters were independent of landscape, i.e., they had a high influence on the absolute value of pollen beetle density but a weak influence on the variation in these densities across maps.
Box plots of the coefficients of the linear model comparing observed and simulated data for a pollen beetle densities and b parasitism rates as a function of the values of selected biological parameters. n = 250 (the total number of simulations per set of 35 maps divided by the five levels). The three parameters were chosen according to the sensitivity indexes on the linear model. Rectangles in grey indicate the range of coefficient values considered acceptable for selecting the best linear model, i.e. that are non-significantly different from 1 and 0 for the slope and intercept, respectively, and that are higher than 20 % for the R². The dashed lines indicate the slope, intercept, and R2 of the best linear model. Each boxplot contains the lower whisker, the lower hinge (first quartile), the median, the upper hinge (third quartile) and the extreme of the upper whisker
A total of 23 parameterizations were selected on the basis of a significance of the linear model, the non-significance of slope and intercept in comparison to 1 and 0, respectively, and a R2 value higher than 0.2. Table 1 and Fig. 4 show parameterizations that most improved the prediction of observations by Mosaic-Pest on the basis of the R2 of the linear regression between observed and simulated data for pollen beetle density (n = 35, Y = 1.05 X + 2.4, R2 = 0.44, P < 0.001) and for parasitism rate (n = 35, Y = 0.8 X + 2.2, R2 = 0.25, P = 0.03). The root mean squared error of standard deviation was 90 and 15 for pollen beetle density and parasitism rates, respectively.
Effect of landscape composition on the influence of biological parameters and identification of the landscape characteristics involved (stage 4 and stage 5)
For pollen beetle density, the value of Morris’ μ* for demographic parameters was highly correlated across maps (r > 0.8, P < 0.05 in all cases). The influence of dispersal parameters such as τMa and βMa were negatively correlated (r < −0.6, P < 0.05) across landscape maps. The influence of demographic parameters was not correlated with dispersal parameters, except that the influence of N Ma0 was related to the influence of ωMa,adult (r = 0.64, P < 0.05).
Concerning the link between the influence of biological parameters and landscape metrics computed at different distances around the central field, the response to landscape metrics differed between one group of demographic parameters (N0, π, and δ) and one group of dispersal parameters (ω, τ, and β) but the response of each parameter within one group was the same. In Fig. 5, we present two examples of parameters, one (π) belonging to the group of demographic parameters and the other (τ) belonging to the group of dispersal parameters. The influence of π was positively related to the proportion of woodlands (r > 0.6, P < 0.05 in all cases) but only for buffer radii <1,000 m (Fig. 5a). The influence of τ was negatively related to the proportion of oilseed rape and of previous oilseed rape in the landscape (r < −0.6, P < 0.05 in all cases) and positively related to the proportion of grasslands and woodlands in the landscape (r > 0.6, P < 0.05 in all cases) for buffer radii >750 m (Fig. 5b).
Spearman’s rank correlation coefficient between the values for the Morris μ* and proportion of habitats between maps across buffer radii. Example of results for a Emergence and b FlightSurvival parameter of pollen beetles. Stars correspond to a Spearman’s rank coefficient that is significantly different from 0 (P < 0.05)
Given the large number of interacting parameters for parasitism rates, inertia analyses on the basis of PCAs were used rather than multiple Spearman’s rank correlations to understand the relationships between groups of parameters and landscape composition. The first factorial plane of the PCA on the values of Morris’ μ* accounted for 78.1 % of the total inertia. This indicates that the influences of demographic parameters of immature pollen beetles and parasitoids were highly correlated across landscape maps (r > 0.8, P < 0.001 in all cases), except for δ Ma, l2 of the pollen beetle and θTh,immature and δ Th,immature of parasitoids, which were correlated in a different group (r > 0.6, P > 0.05) (Fig. 6a). Dispersal parameters were also correlated in two distinct groups, one corresponding to pollen beetle parameters and one to parasitoid parameters (r > 0.6, P < 0.001 in all cases) (Fig. 6a).
Co-inertia analysis was significant (P = 0.005, based on 999 repetitions of Monte-Carlo simulations). The co-inertia analysis of the PCA of the values of Morris’ μ* projected on the PCA of the Landscape metrics data set explained 76.3 % of the variability (Fig. 6a). The first factorial plane of the PCA on the Landscape metrics data set accounted for 59.2 % of the total inertia (Fig. 6b). Not surprisingly, landscape metrics describing the proportion of a given habitat at several buffer sizes were correlated among themselves. For the relationships between the Landscape metrics data set and the values of Morris’ μ*, it appeared that dispersal parameters of parasitoids (r > 0.6, P < 0.05) were related to grasslands whereas demographic parameters of pollen beetles were related to woodlands (r > 0.6, P < 0.05), except for three parameters (δ Ma,l2 of pollen beetle and θTh,immature and δ Th,immature of parasitoids) (Fig. 6a, b). Two dispersal parameters of pollen beetles, ωMa,adult and βMa, were linked to the proportion of previous oilseed rape fields at distances <1,000 m and to the proportion of oilseed rape fields at distances up to 750 m, respectively (r > 0.6, P < 0.01). Considering multicollinearities between buffers of a given habitat, no important distance effect was evident in the relationship between semi-natural habitats (woodlands and grasslands) and biological parameters.
Discussion
The Mosaic-Pest model integrates all the existing knowledge about the ecology of M. aeneus and T. heterocerus with regard to landscape effects. When studying the relationships between simulated and observed values (Fig. 1, stage 2), we demonstrated that Mosaic-Pest is able to reproduce general trends of the system and of pollen beetle dynamics. The relationship between simulated and observed values was weaker for parasitism rates than for pollen beetle dynamics, and we therefore suspect that important processes need to be added in the model. As expected, the influences of dispersal parameters on model accuracy and precision were highly variable between the studied landscape maps because landscape composition affects distances between complementary and/or supplementary habitats. The effects of demographic parameters were also highly complex when they corresponded to processes occurring after dispersal events, such as duration of adult’s stage.
The sensitivity analysis of the Mosaic-Pest model highlights the importance of correctly specifying key parameters in studying the spatio-temporal dynamics of the pollen beetle. Among the 32 studied parameters, six and 19 greatly affect pollen beetle densities and parasitism rates, respectively. For example, the dispersal event before feeding is crucial and relies on parameters that should be re-examined. Moreover, the proportion of adults able to emerge after overwintering is also a key parameter but is rather difficult to estimate despite recent studies on the overwintering period (Nilsson 2010). We found a higher number of influential parameters for parasitism rates than for density of pollen beetles because the former involved more processes. Moreover, the large size of box plots indicates that the value for Morris μ* was highly variable across landscape maps, which points out the importance of using several landscapes varying in habitat composition for sensitivity and validation procedures: the importance of parameters is greatly influenced by the landscape.
The use of multivariate analyses helps explain the influence of dispersal parameters in the system and their strong link with landscape composition. For pollen beetle densities, both proportion of individuals that survive per meter travelled and the decrease in habitat attractiveness with distance travelled depended on the distance covered during dispersal. The influence of the proportion of individuals that survive per meter travelled was negatively related to the proportion of oilseed rape and previous oilseed rape in the landscape and was positively related to the proportion of grasslands and woodlands within buffer radii >750 m. Two other dispersal parameters of pollen beetles, the maximum dispersal distance travelled by insects before feeding and the decrease in habitat attractiveness with distance travelled, were also linked to the proportion of previous oilseed rape fields. The results of co-inertia analysis suggest that an increase in the proportion of oilseed rape fields enhances the connectivity between fields and limits the importance of survival during dispersal. For example, the co-inertia analysis highlighted that dispersal parameters of parasitoids were related to grasslands, whereas demographic parameters of pollen beetles were related to woodlands. The effect of grasslands is reasonable because grasslands enhance the spread of parasitoids in the landscape and consequently enhance parasitism rates. The influence of demographic parameters is related to woodlands because these habitats act as sources of pollen beetles, i.e., as hosts for parasitoids. This provision of resources is more important when distances are small.
These results indicate that dispersal parameters of parasitoids before feeding are crucial and that their estimates must be improved. This is especially the case for the quantitative link between the dispersal window of parasitoids and the proportion of grasslands in the landscape. Obtaining better estimates of how landscape affects parasitoid dispersal parameters should enhance the implementation of innovative pest management practices that affect demographic traits of pests and natural enemies because the efficiency of these practices will vary with landscape composition and structure, as pointed out by Tscharntke et al. (2005). The results on the relationship between parameter influence and landscape composition will help in identifying the landscapes in which a given biological parameter, e.g., density of overwintering insects or dispersal, will be the most important and will therefore enable researchers to focus on situations where the parameter is expected to show the greatest influence. For example, the Mosaic-Pest model will help researchers select contrasting landscapes for calibrating dispersal processes. From a more applied perspective, knowledge of the relationship between landscape and parameter influence will facilitate the selection of a pest management strategy that focuses on a vulnerable part of the pest life cycle in a given landscape.
Studying the interaction between landscape composition and the influences of parameters on the output of a spatial model is novel and is important for understanding processes in landscape ecology. The co-inertia analysis helps us understand how the main effects of the model parameters on model outputs were affected by landscape composition. Co-inertia analyses are particularly suitable for studying ecological data tables in a symmetric way (ter Braak and Schaffers 2004); co-inertia analysis, for example, has been used to study the relationship between species composition and environmental variables sampled at the same location (Dray et al. 2003). We extended the use of co-inertia analyses to disentangle the complex links between landscape composition and ecological processes via a spatially explicit model. We hope that the new methodology presented here will help landscape modellers to go deeper in the understanding of the link between their model and landscape configuration.
The approach presented in our study, which combines Morris sensitivity analysis and variance decomposition techniques, provided useful results for our case study and could be part of the toolbox of inverse modelling techniques. The use of a given inverse modelling technique for calibrating model parameters is widely dependent on model computational time and number of parameters to assess. For example, Bayesian techniques (Martinez et al. 2011) or genetic algorithm (Stonedahl and Wilensky 2010) could be tested with success on some spatially explicit models with a low number of parameters. Global sensitivity analyses are the best techniques to calculate a synthetic measure of parameters influence, the parameters space uncertainty being explored via Morris method as presented in this paper, or latin hypercube sampling (Luxmoore et al. 1991) and a complete factorial plan (Beaudouin et al. 2008), depending on number of parameters evolved. For researchers interested in linking parameter influence to landscape metrics, different type of multivariate analyses could be found, depending as well on the number and the type of response variable to link (a complete review of the multivariate analyses could be find in Ramette 2007).
References
Beaudouin R, Monod G, Ginot V (2008) Selecting parameters for calibration via sensitivity analysis: an individual-based model of mosquito fish population dynamics. Ecol Model 218:29–48
Bianchi FJJA, Schellhorn NA (2009) Foraging behaviour of predators in heterogeneous landscapes: the role of perceptual ability and diet breadth. Oikos 118:1363–1372
Cariboni J, Gatelli D, Liska R, Saltelli A (2007) The role of sensitivity analysis in ecological modelling. Ecol Model 203:167–182
Charnell MA (2008) An individual-based model of a tritrophic ecology. Ecol Model 218:195–206
Cook S, Murray D, Williams I (2004) Do pollen beetles need pollen? The effect of pollen on oviposition, survival, and development of a flower-feeding herbivore. Ecol Entomol 29:164–173
Cook SM, Khan ZR, Pickett JA (2007) The use of push-pull strategies in integrated pest management. Annu Rev Entomol 52:375–400
Csilléry K, Blum MGB, Gaggiotti OE, François O (2010) Approximate Bayesian computation (ABC) in practice. Trends Ecol Evol 25:410–418
Dray S, Chessel D, Thioulouse J (2003) Co-inertia analysis and the linking of ecological data tables. Ecology 84:3078–3089
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Frair JL, Merrill EH, Visscher DR, Fortin D, Beyer HL, Morales JM (2005) Scales of movement by elk (Cervus elaphus) in response to heterogeneity in forage resources and predation risk. Landscape Ecol 20:273–287
Grimm V, Revilla E, Berger U, Jeltsch F, Mooij WM, Railsback SF, Thulke HH, Weiner J, Wiegand T, DeAngelis DL (2005) Pattern-oriented modeling of agent-based complex systems: lessons from ecology. Science 310:987–991
Hartig F, Calabrese JM, Reineking B, Wiegand T, Huth A (2011) Statistical inference for stochastic simulation models—theory and application. Ecol Lett 14:816–827
Herrström G (1964) Untersuchungen über Parasiten von Ölfruchtschädlingen in Sweden. Meddn StVäxtskAnst 12:433–448
Jourdheuil P (1960) Influence de quelques facteurs écologiques sur les fluctuations de population d’une biocénose parasitaire. INRA, Paris
Kramer-Schadt S, Revilla E, Wiegand T, Breitenmoser U (2004) Fragmented landscapes, road mortality and patch connectivity: modelling influences on the dispersal of Eurasian lynx. J Appl Ecol 41:711–723
Kramer-Schadt S, Revilla E, Wiegand T, Grimm V (2007) Patterns for parameters in simulation models. Ecol Model 204:553–556
Luxmoore RJ, King AW, Tharp ML (1991) Approaches to scaling up physiologically based soil–plant models in space and time. Tree Physiol 9:281–292
Martinez I, Wiegand T, Batllori E, Gutierrez E (2011) Disentangling the formation of contrasting tree-line physiognomies combining model selection and Bayesian parameterization for simulation models. Am Nat 177:E136–E152
McIntire EJB, Fajardo A (2009) Beyond description: the active and effective way to infer processes from spatial patterns. Ecology 90:46–56
Mills NJ, Getz WM (1996) Modelling the biological control of insect pests: a review of host–parasitoid models. Ecol Model 92:121–143
Monod H, Naud C, Makowski D (2006) Uncertainty and sensitivity analysis for crop models. In: Wallach D, Makowski D, Jones J (eds) Working with dynamic crop models. Elsevier, Amsterdam
Morris MD (1991) Factorial sampling plans for preliminary computational experiments. Technometrics 33:161–174
Nilsson C (1988a) The pollen beetle (Meligethes aeneus F.) in winter and spring rape at Alnarp 1976–1978. I. Migration and sex ratio. Växtskyddsnotiser 56:6
Nilsson C (1988b) The pollen beetle (Meligethes aeneus F.) in winter and spring rape at Alnarp 1976–1978. II. Oviposition. Växtskyddsnotiser 52: 139–144
Nilsson C (2010) Impact of soil tillage on parasitoids of oilseed rape pests. In: Williams I (ed) Biocontrol-based integrated management of oilseed rape pests. Springer, Dordrecht, pp 45–76
R Development Core Team (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160
Ricci B, Franck P, Toubon JF, Bouvier JC, Sauphanor B, Lavigne C (2009) The influence of landscape on insect pest dynamics: a case study in southeastern France. Landscape Ecol 24:337–349
Rusch A, Valantin-Morison M, Sarthou J-P, Roger-Estrade J (2011) Multi-scale effects of landscape complexity and crop management on pollen beetle parasitism rate. Landscape Ecol 26:473–486
Saltelli A, Tarantola S, Campolongo F (2000) Sensitivity analysis as an ingredient of modeling. Stat Sci 15:377–395
Stonedahl F, Wilensky U (2010) Evolutionary robustness checking in the artificial Anasazi model. In: Proceedings of the AAAI fall symposium on complex adaptive systems: resilience, robustness, and evolvability. Arlington, VA, pp 120–129
Taimr L, Sedivy J, Bergmannova E, Hanker I (1967) Further experience obtained in studies on dispersal flights of Meligethes aeneus F., marked with P32 (Coleoptera). Acta Entomol Bohemoslov 64:325–332
ter Braak CJF, Schaffers AP (2004) Co-correspondence analysis: a new ordination method to relate two community compositions. Ecology 85:834–846
Thiele JC, Grimm V (2010) NetLogo meets R: linking agent-based models with a toolbox for their analysis. Environ Model Softw 25:972–974
Tscharntke T, Brandl R (2004) Plant–insect interactions in fragmented landscapes. Annu Rev Entomol 49:405–430
Tscharntke T, Klein AM, Kruess A, Steffan-Dewenter I, Thies C (2005) Landscape perspectives on agricultural intensification and biodiversity—ecosystem service management. Ecol Lett 8:857–874
Viaud V, Monod H, Lavigne C, Angevin F, Adamczyk K (2008) Spatial sensitivity of maize gene-flow to landscape pattern: a simulation approach. Landscape Ecol 23:1067–1079
Vinatier F, Gosme M, Valantin-Morison M (2012a) A tool for testing integrated pest management strategies on a tritrophic system involving pollen beetle, its parasitoid and oilseed rape at the landscape scale. Landscape Ecol 27:1421–1433
Vinatier F, Lescourret F, Duyck P-F, Tixier P (2012b) From IBM to IPM: using individual-based models to design the spatial arrangement of traps and crops in integrated pest management strategies. Agric Ecosyst Environ 146:52–59
Wiegand T, Moloney KA, Naves J, Knauer F (1999) Finding the missing link between landscape structure and population dynamics: a spatially explicit perspective. Am Nat 154:605–627
Wiegand T, Knauer F, Kaczensky P, Naves J (2004) Expansion of brown bears (Ursus arctos) into the eastern Alps: a spatially explicit population model. Biodivers Conserv 13:79–114
Wiegand T, Revilla E, Moloney KA (2005) Effects of habitat loss and fragmentation on population dynamics. Conserv Biol 19:108–121
Wilensky U (1999) Netlogo. Center for Connected Learning and Computer-Based Modeling, Evanston
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vinatier, F., Gosme, M. & Valantin-Morison, M. Explaining host–parasitoid interactions at the landscape scale: a new approach for calibration and sensitivity analysis of complex spatio-temporal models. Landscape Ecol 28, 217–231 (2013). https://doi.org/10.1007/s10980-012-9822-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-012-9822-4