Abstract
We utilize empirically derived estimates of landscape resistance to assess current landscape connectivity of American marten (Martes americana) in the northern Rocky Mountains, USA, and project how a warming climate may affect landscape resistance and population connectivity in the future. We evaluate the influences of five potential future temperature scenarios involving different degrees of warming. We use resistant kernel dispersal models to assess population connectivity based on full occupancy of suitable habitat in each of these hypothetical future resistance layers. We use the CDPOP model to simulate gene exchange among individual martens in each of these hypothetical future climates. We evaluate: (1) changes in the extent, connectivity and pattern of marten habitat, (2) changes in allelic richness and expected heterozygosity, and (3) changes in the range of significant positive genetic correlation within the northern Idaho marten population under each future scenario. We found that even moderate warming scenarios resulted in very large reductions in population connectivity. Calculation of genetic correlograms for each scenario indicates that climate driven changes in landscape connectivity results in decreasing range of genetic correlation, indicating more isolated and smaller genetic neighborhoods. These, in turn, resulted in substantial loss of allelic richness and reductions in expected heterozygosity. In the U.S. northern Rocky Mountains, climate change may extensively fragment marten populations to a degree that strongly reduces genetic diversity. Our results demonstrate that for species, such as the American marten, whose population connectivity is highly tied to climatic gradients, expected climate change can result in profound changes in the extent, pattern, connectivity and gene flow of populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Changes in climate affect a range of ecological conditions, leading to change in species distributions and range shifts. There has been a proliferation in research about the potential effects of climate change on range shifts and population connectivity. Many researchers have utilized the “bioclimatic envelope” approach, which predicts future shifts in the climatic conditions currently associated with species distributions (Iverson et al. 1999; McKenzie et al. 2003; Rehfeldt et al. 2006). There are several major limitations of this approach. Most importantly, they project spatial shifts in niche-suitability but do not typically explicitly address the ability of the focal species to disperse from current to projected future ranges. Without explicit assessment of species migration potential, bioclimatic niche projections merely show the changes in the distribution of suitable environmental conditions, which may be very different than likely future distributions if the species has limited dispersal ability or if substantial landscape barriers lie between current and projected future ranges. Second, most past work on projecting climate driven changes in species distributions are conducted at coarse spatial scales, in which fine-scale environmental variability is averaged away. Such coarse-scale models of environmental control may miss important drivers of ecological variability. The dominant scales of environmental control typically include the scale at which species directly interact with their environment (Cushman et al. 2007, 2010a). Analyses conducted at inappropriate scales may fail to identify potential refugia or accurately assess migration potential and likely dispersal routes (Wiens et al. 2009; Cushman et al. 2010b). Reliable prediction of climate change impacts on species distributions requires empirically based, process-driven analyses conducted at the scales that dominate the species-environment relationship (e.g. Cushman et al. 2007; Evans and Cushman 2009).
Recent research has indicated that population connectivity of American marten (Martes americana) in the U.S. northern Rocky Mountains is controlled by elevational gradients in landscape resistance to gene flow (Wasserman et al. 2010). The optimal resistance model identified in Wasserman et al. (2010) is closely related to areas of high winter snowpack and moist montane forest. We speculate that this strong relationship between gene flow and elevation reflects marten requirement for deep snowpacks (suggesting avoidance of low elevations) in interaction with habitat preference for mesic, middle elevation forest (which suggests avoidance of lowest and highest elevations; Wasserman 2008). The lack of association with forest cover, forest type and other habitat features suggests that movement of dispersing juvenile martens is primarily related to elevation. This, in turn, suggests an association with climate.
Climate change is predicted to result in large increases in winter temperature in the northern Rocky Mountains (IPCC 2007; Littell et al. 2011). This is likely to result in substantial decrease in the depth of average winter snowpacks (Elsner et al. 2010; Littell et al. 2011) and migration of forest communities upward in elevation. Given the biology of the American marten and its association with elevational gradients, this will increase the average resistance of the landscape to martens and could result in increased isolation of remnant populations in high elevation mountains separated by the deep river valleys that bisect the region. This suggests that population connectivity of American martens in the northern Rocky Mountains may be highly vulnerable to climate change.
Objectives
The goal of this study is to predict the effects of future climate change on population connectivity and genetic characteristics of an American marten population in the U.S. northern Rocky Mountains. To accomplish this goal, we address three research objectives. First, we use a landscape resistance map derived from empirical landscape genetic modeling to predict the current extent and fragmentation of American marten dispersal habitat. Second, we use a spatially explicit simulation model to predict the population genetic characteristics of this current population as functions of landscape resistance and compare predictions with the actual current genetic structure of the marten population. Third, we evaluate changes in the extent and fragmentation of marten habitat under five scenarios involving upward shifts of current temperatures by 100, 200, 300, 400, and 500 m, corresponding to climatic warming of between 0.65 and 3.25°C, bracketing the range of temperature increase expected in the region for the middle of the twenty-first century (Mote and Salathé 2010). Fourth, we simulate changes in population genetic structure of the marten population under each of these scenarios.
Methods
Study area
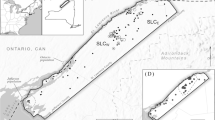
The study area consists of an approximately 5500 km2 area in the Selkirk, Purcell and Cabinet Mountains, including portions of the states of Idaho, Montana, Washington and the province of British Columbia (Fig. 1). Landownership consists of U.S. Forest Service (Idaho Panhandle National Forest), state, Crown, provincial and private timber lands, and private residences. The topography is mountainous, with steep ridges, narrow valleys and many cliffs and cirques at the highest elevations. Elevation ranges from approximately 700 to 2700 m, with upper tree line at approximately 2500 m. The Kootenai River divides the study area, separating the Selkirk Mountains on the west from the Purcell Mountains on the east with a 5–8 km wide unforested, agricultural valley and a broad, deep river. Other major drainages include the Pend Oreille River, on the far western edge of the study area, and the Priest River in the western third. The area is heavily forested, with Abies lasiocarpa (subalpine fir) and Picea engelmannii (Engelmann spruce) codominant above 1300 m, and a diverse mixed conifer forest dominating below 1300 m. For further description of the vegetation community in the study area see Cushman et al. (2006) and Evans and Cushman (2009).
Study area orientation map and current landscape resistance. The study area includes approximately 5500 km2 in northern Idaho, northeastern Washington and northwestern Montana, as well as southern British Columbia. The map in the top panel depicts the resistance map produced by Wasserman et al. (2010)
Landscape resistance model
We utilize a landscape resistance map for American marten derived from an empirical landscape genetic analysis (Wasserman et al. 2010). The analysis used a univariate scaling analysis within a two-step form of the causal modeling approach (Cushman et al. 2006) to integrate model selection with null hypothesis testing in individual-based landscape genetic analysis. The causal modeling approach to landscape genetic inference has been shown to have high power to identify the correct generating process and reject spurious correlated alternative hypotheses (Cushman and Landguth 2010). Wasserman et al. (2010) found that gene flow in American marten in northern Idaho is primarily related to elevation, and that alternative hypotheses involving isolation by distance, geographical barriers, effects of canopy closure, roads, tree size class and an empirical habitat model are not supported.
Gene flow in the Northern Idaho American marten population is driven by an elevational gradient of landscape resistance. Landscape resistance to gene flow is at minimum at 1500 m and resistance increases as a Gaussian function of elevation with a 300 m standard deviation to a maximum resistance of 10 (Fig. 1). The resistance value in the cost map represents the cost of crossing each cell relative to the least-cost landscape condition (1500 m). This resistance layer is used as the basis for all analyses of current population connectivity and simulation of current landscape genetic structure. It is also used as the starting point for predicting future landscape resistance under the five climate change scenarios.
Climate change scenarios
Projected climate changes in the Pacific Northwest vary with global climate model, time frame, season and greenhouse gas emissions scenario. Mote and Salathé (2010) project increases in Pacific Northwest (PNW) regional autumn, winter, and spring temperature of about 1.4–2.0°C by the 2040s and 2.1–3.4°C by the 2080s (relative to 1970–1999, A1B and B1 emissions scenarios average across 20 and 19 IPCC AR4 global climate models (GCMs), respectively). Extending methods described in Elsner et al. (2010), Littell et al. (2011) and McKelvey et al. (in press) used a basic spatially explicit downscaling method to downscale monthly PNW regional projections from 10 best-ranked GCMs to 1/16th degree (~5 × 7 km) and reported projected climate changes by ecosections. For the ecosection containing the landscape in this study (Bailey M333A), the 10 GCM ensemble cool season (October–March) temperature changes are expected to average +1.7°C by the 2040s (2030–2059) and +3.3°C by the 2080s (2070–2099) relative to the 1970–1999 mean of −0.4°C.
Assuming a long term average standard environmental lapse rate (rate of decrease in temperature with elevation) of −6.5°C/km, this is equivalent to an increase in the elevation of current temperatures of approximately 270 m by the 2040s and 500 m by the 2080s (relative to 1970–1999). While lapse rates vary considerably with topography and season in the Pacific Northwest (e.g., Minder et al. 2010), the standard environmental lapse provides a baseline for future comparison. More importantly, this temperature increase crosses the 0°C isotherm for the landscape (historical average October–March mean temperature estimated to be −0.4°C for ecosection M333A), suggesting a decline in the proportion of winter precipitation falling as snow. Averaged over the M333A ecosection, snowpack is projected to change −75% by the 2040s under the 10 GCM ensemble and an A1B emissions scenario (Littell et al. 2011).
We specified five scenarios of climate driven changes to landscape resistance to American marten in the study area. Specifically, we evaluated the effects of upward shift of current temperatures by 100, 200, 300, 400, and 500 m from the current optimum elevation of 1500 m (Wasserman et al. 2010). This represents a rather conservative range of scenarios, with an upward shift of mean annual temperatures of approximately 500 m expected by 2080 (Littell et al. 2011). Matching our scenarios with Littell et al. (2011) our +300 m scenario approximates expected climatic warming for year 2040, and our +500 m approximate warming expected by 2080.
Modeling current and future habitat connectivity
We predicted current and future habitat connectivity for American marten using a least-cost resistant kernel approach (e.g. Compton et al. 2007; Cushman et al. 2010a, b) coupled with landscape pattern analysis with FRAGSTATS (McGarigal et al. 2002). The resistant kernel approach to modeling landscape connectivity has a number of advantages as a robust approach to assessing current population connectivity for multiple wildlife species under climate change scenarios. First, unlike most corridor prediction efforts, it provides prediction and mapping of expected migration rates for every pixel in the study area, rather than only for a few selected “linkage zones” or narrow corridors (e.g. Compton et al. 2007). Second, scale dependency of dispersal ability can be directly included to assess how species of different vagilities will be affected by landscape change and fragmentation under a range of scenarios (e.g. Cushman et al. 2010a). Third, it is computationally efficient, enabling simulation and mapping at a fine spatial scale across large geographical extents (e.g. Cushman et al. 2010b, 2011a).
The resistant kernel approach to connectivity modeling is based on least-cost dispersal from a defined set of sources. The sources in our case are a uniform grid of pixels seeded onto the resistance maps. The source grid places dispersing individuals every 1.5 km in cells with landscape resistance less than 2. This roughly reflects the spacing of home ranges for this species in this landscape (Wasserman 2008), and restricts origin cells to locations of relatively high suitability as dispersal habitat for the species (Wasserman et al. 2010). Dispersal habitat is different than home-range habitat relationships (Wasserman 2008; Wasserman et al. 2010). The model employed in this paper predicts genetic differentiation (and is based purely on elevation; Wasserman et al. 2010), while habitat utilization (Cushman 2011b; Wasserman 2008) of marten in this study area is related to a more complex suite of factors (elevation, roads, forest type, patch density, seral stage). As the goal of this paper is to predict population connectivity we limit analyses to the resistance map based on dispersal (Wasserman et al. 2010). The costs reflected in the resistance map are used as weights in the dispersal function, such that the expected population density in a pixel is down-weighted by the cumulative cost from the source, following the least-cost route (Compton et al. 2007).
The model calculates the expected relative density of American marten in each pixel around the source, given the dispersal ability of the species, the nature of the dispersal function, and the resistance of the landscape (Compton et al. 2007; Cushman et al. 2010a). We wrote an ESRI ArcGrid script to calculate the resistant kernel (R k) density (ESRI 2005). The script uses the ArcGrid COSTDISTANCE function to produce a map of the movement cost from each source up to a specified dispersal threshold. These cost distance grids are inverted and scaled such that the maximum value for each individual kernel is one. Once the expected density around each source cell is calculated, the kernels surrounding all sources are summed to give the total expected density at each pixel. The results of the model are surfaces of expected density of dispersing organisms at any location in the landscape.
We specified a dispersal threshold of 15,000 cost units, reflecting a dispersal ability of 15 km in uniformly suitable habitat. Dispersal is poorly understood in American marten due to the limited number of studies using radio telemetry to monitor dispersal events. Most such studies are limited by very small sample sizes making generalization difficult. Bull and Heater (2001) report an average dispersal distance of 18.6 km in an Oregon study area. Broquet et al. (2006) note that marten populations reintroduced into areas from which they had previously been extirpated typically expand slowly at rates of less than 16 km per decade (de Vos 1951; Gardner and Gustafson 2004), suggesting relatively limited dispersal. Average dispersal distance is typically correlated with home range size, with longer average dispersal in systems with larger than average home ranges. Our study area has home ranges that are much smaller on than average across the species range (Tomson 1999). Therefore we expect relatively modest average dispersal distances. Based on these studies, we believed that a 15 km dispersal threshold reflects a reasonable value. The resistant kernel models produce maps of expected connected habitat for American marten in each resistance landscape, assuming a 15 km dispersal threshold.
Analysis of kernel connectivity maps
To quantify the extent and connectivity of connected American marten dispersal habitat, the maps were reclassified into binary form for analysis in FRAGSTATS. Any cell with non-zero kernel density value was classified as connected and all pixels with zero kernel density were classified as non-connected. We used FRAGSTATS (McGarigal et al. 2002) to calculate the percentage of the landscape, largest patch index and number of patches of predicted dispersal habitat in the resistance maps for the current and five future climate change scenarios. The percentage of the landscape is the simplest metric of landscape composition, and quantifies how much of the study area is covered by potentially occupied habitat for each species and species group. The percentage of the landscape, however, does not quantify any information about the configuration of that potential habitat. Population connectivity is a function of the ability of organisms to traverse continuously through connected habitat. We calculated two additional landscape metrics to provide a view of the degree of connectivity and fragmentation of these core habitats. The number of patches of connected habitat in the landscape provides an intuitive measure of the degree of fragmentation of the landscape for marten dispersal. When habitat is fully connected there is a single patch, and as habitat becomes progressively fragmented habitat is broken up into successively more separate isolated patches. It gives a global measure of the connectivity of dispersal habitat in the study area and is a more relevant functional measure than are patch size, nearest neighbor distance and percentage of the landscape in occupied habitat (McGarigal et al. 2002). Third, we calculated the largest patch index (McGarigal et al. 2002) of connected core habitat. This index reports the extent, as a proportion of the size of the study area, of the largest patch of connected core habitat.
Identifying barriers and fracture zones
The FRAGSTATS analyses described above provide quantitative evaluation of the extent and fragmentation of dispersal habitat for American marten in the study area. However, to be most useful to guide landscape planning and climate adaptation, analyses need to provide spatially explicit predictions of the location of barriers breaking up populations and of fracture zones where connectivity is reduced. Mapping barriers and fracture zones provides the information needed for managers to identify the most critical locations in the landscape for conservation or restoration. We identified barriers as locations where the resistant kernel predictions became zero between patches of core habitat. We identified fracture zones as locations where the resistant kernel predictions resulted in narrow isthmuses joining larger blocks of connected dispersal habitat.
Simulating current and future population genetic structure
Simulation modeling provides explicit control over pattern-process relationships (Bruggeman et al. 2010; Epperson et al. 2010). This enables rigorous attribution of the causes of observed differentiation, which is not possible in empirical studies. Simulation models offer a flexible way to investigate the behavior of idealized ecological processes in idealized landscapes. By varying functional parameters, environmental characterization, and organism attributes, scientists can investigate hypotheses about the relative influence of different factors, their interactions, and ranges of organism characteristics, such as gradients of landscape resistance, population size or dispersal ability. This provides a means for thorough evaluation of complexes of factors that would be impossible to investigate directly in the field. In addition, simulations are particularly important in evaluating scenarios of future landscape change, such as the effects of climatic warming on landscape resistance and its effects on future population genetic structure.
We used CDPOP 0.84 (Landguth and Cushman 2010) to simulate the processes of mating and dispersal as functions of the spatial patterns of habitat and non-habitat on the six resistance maps corresponding to the current landscape and five scenarios of future landscape resistance. CDPOP is an individual-based, spatially explicit, landscape genetic program that simulates birth, death, mating and dispersal of individuals in complex landscapes as probabilistic functions of movement cost among them. In each of the six landscape maps, we used the same source cells described above in the resistant kernel analysis as locations of individual martens. We simulated gene flow among these locations for 500 non-overlapping generations. We stipulated the population to have 30 loci, with 10 alleles per locus, initially randomly assigned among individuals and a mutation rate of 0.0005. We used an inverse square mating and dispersal probability function, with maximum dispersal cost-weighted distance of 15000 m (the same dispersal threshold used in the resistant kernel analyses described above). Reproduction was sexual with non-overlapping generations, and the number of offspring was based on a Poisson probability draw with mean of 4. For each of the six resistance scenarios, we ran 25 Monte Carlo runs in CDPOP to assess stochastic variability.
Assessing local and global population structure
We ran all CDPOP simulations for 500 generations. Past studies have shown that this is sufficient time to ensure spatial genetic equilibrium (Landguth et al. 2010a, b). We extracted several global measures of population genetic structure for the full study area at generation 500. These include total number of alleles in the population and expected heterozygosity. We analyzed the differences in these global measures of genetic structure between the six different resistance scenarios. In addition, calculated Pearson correlations between these measures of genetic structure and the FRAGSTATS metrics to explore the relationship between these genetic attributes and landscape patterns of connected habitat, and determine if some genetic attributes are dominantly related to habitat area while others are primarily related to habitat fragmentation.
We also assessed local population genetic structure across all six resistance scenarios. We utilized the sGD program (Shirk and Cushman 2011) to map local total number of alleles and expected heterozygosity. sGD uses a variable radius focal window to calculate locally centered values of a number of population genetic parameters, such as allelic richness, expected and observed heterozygosity, across spatially structured populations. We calculated the local number of alleles and expected heterozygosity within local windows with width equal to 1/2 of the range of significant spatial genetic autocorrelation of the Mantel correlogram in the actual marten population in the study area (Wasserman et al. 2010). This provides spatially explicit evaluation of the population genetic consequences of the different barriers and fracture zones identified by the process described above, and a means to spatially evaluate the pattern and magnitude of climate change effects on population genetic structure across the study area.
Predicting effects of climate change on genetic neighborhood extent
We compared Mantel correlograms for the simulated current and five future landscape resistance scenarios to determine whether habitat fragmentation resulting from climate change is likely to alter the extent of the genetic neighborhoods of American marten in the study area. A shortening of the range of significant genetic autocorrelation as a function of cost distance would indicate that simulated climate change fragments marten populations and reduces the spatial extent of genetic neighborhoods.
Results
Resistant kernel modeling of connected dispersal habitat
Maps showing predicted connected dispersal habitat for all six resistance scenarios are shown in Fig. 2. In all scenarios populations were predicted to be restricted to mountain ranges separated by low elevation barriers, such as the Kootenai, Priest and Pend Oreille Rivers. Under the current resistance condition, dispersal habitat for American marten was predicted to exist in several large blocks (Fig. 2a). Notably, nearly all dispersal habitat in the Selkirk Range was predicted to exist in a single large connected patch. Likewise, all dispersal habitat in the Cabinet Range was predicted to reside in a single connected patch. In contrast, the Purcell Range was predicted to be broken into seven distinct patches that are beyond the simulated dispersal distance from any other patches of dispersal habitat.
Resistant kernel maps of predicted connected dispersal habitat for American marten under six scenarios of climate change. a Current population connectivity using Wasserman et al. (2010) as the resistance map in the kernel model (optimum 1500 m), and upward movement of optimum resistance elevation by b +100 m, c +200 m, d +300 m, e +400 m, and f +500 m. In the maps, black areas are predicted to have non-zero positive kernel density, indicating they are part of connected patches of dispersal habitat, while gray cells have zero kernel density, indicating they are outside any connected habitat
Habitat was reduced and fragmented across the five climate change scenarios (Fig. 2b–f). In the +200 m scenario, the Purcell populations were predicted to be highly fragmented, with several currently connected large populations broken up (Fig. 2d). However, the large and connected dispersal habitat patches in the Selkirk and Cabinet Ranges remained intact. However, in the +400 m scenario, two fracture zones appeared in the Selkirk Range, threatening connectivity between the American and Canadian portions of the range (Fig. 2e). Similarly, at +500 m a fracture zone appeared in the Cabinet Range, which may represent diminished connectivity from the northern and southern parts of that range within the study area. At +500 m there was extensive habitat loss and fragmentation (Fig. 2f). In the Purcell and Cabinet Ranges, connected habitat was predicted to be reduced to a number of small and isolated remnant patches. Predicted habitat loss and fragmentation was less severe in the Selkirk Range. However, the resistant kernel model at scenario +500 m predicted functional isolation of the Canadian and American portions of the range, with several barriers predicted along Boundary and Smith Creeks which isolate the Canadian from the American patches of connected dispersal habitat.
FRAGSTATS analysis of habitat loss and fragmentation resulting from simulated climate change
All three FRAGSTATS metrics indicated substantial effects of climate change on landscape connectivity (Fig. 3). The percentage of the landscape occupied by connected dispersal habitat decreased from approximately 65–27% across the scenarios. This indicated a roughly 50% loss in predicted connected habitat for the species between current and the +500 m scenario, reflecting expected climatic warming at year 2080. The percentage of the landscape occupied by dispersal habitat decreased monotonically and roughly linearly, indicating there no thresholds in the effect of climate change on the area of dispersal habitat for American marten.
Graph of three FRAGSTATS metrics (percentage of the landscape occupied by connected dispersal habitat (PLAND), percentage of the landscape in the largest patch of connected habitat (LPI), and the number of isolated patches of connected habitat (NP) across the six resistance scenarios reflecting optimum dispersal elevations of 1500, 1600, 1700, 1800, 1900 and 2000 m
In contrast, the two configuration metrics, largest patch index (LPI) and number of patches (NP) both showed threshold relationships. Both NP and LPI were relatively stable across scenarios current to +300 m. However, they changed rapidly between +300 and +500 m, with the number of isolated patches increasing rapidly and the size of the largest remaining connected patch decreasing rapidly. From scenario +300 to +500 m the number of isolated patches of marten habitat increased by nearly 80% and the largest patch index decreased by nearly 2/3. In contrast to the linear decline in the area of connected habitat, these results show that fragmentation of the landscape into isolated patches occured suddenly in a threshold-like phenomenon when the optimum elevation for marten dispersal exceeds 1800 m (scenario +300 m).
Effects of climate change on global population structure
Number of alleles in the population decreased monotonically and roughly linearly across the scenarios (Fig. 4). Expected heterozygosity appeared to follow a curvilinear response to increasing temperature, with relatively little decrease up to scenario +300 m, followed by a much more rapid decrease (Fig. 5).
Effects of climate change on local population structure
In the current landscape, allelic richness was highest in the central Selkirk and Purcell mountains (Fig. 6a). Allelic richness declined dramatically across the climate change scenarios (Fig. 6b–e) in all parts of the study area, with the highest allelic diversity predicted under each scenario in the central portion of the Selkirk Mountains. A very similar pattern was seen for local expected heterozygosity (data not shown). As the population is fragmented, local expected heterozygosity decreased, such that by scenario +400 m local expected heterozygosity had declined dramatically across the population (Fig. 5).
Relationship between landscape structure and population structure
The number of alleles in the population and the expected heterozygosity were both very highly correlated with PLAND and LPI, while expected heterozygosity also highly correlated with the number of isolated patches of connected habitat (Table 1).
Effect of climate change on extent of genetic neighborhoods
The current marten population in the study area is expected to have relatively broad-scale genetic relatedness (Fig. 7). Specifically, the range of significant correlation of genetic distances among individual martens in our simulation approximated 92,000 cost units in the current resistance landscape (Fig. 7a). This is very similar to the range of the empirical correlogram of actual genetic data produced by Wasserman (2008), which has a range of approximately 83,000 cost units. The range of significant genetic correlation decreased monotonically, and variance increased, across all five climate change scenarios (Fig. 7b–f). This indicates that habitat loss and fragmentation due to climate change may result in smaller and more isolated genetic neighborhoods. This reduction in the extent of correlated genetic neighborhoods was predicted to be quite large. For example, the range of significant genetic correlation in the +500 m scenario was roughly 50000 cost units, equivalent to a 46% decrease the spatial extent of that simulated in the current landscape (Fig. 7f).
Mantel correlograms showing the multivariate genetic correlation between individuals separated by successive lag distance classes for each of the six scenarios. The scenarios are optimum movement habitat at a 1500 m, b 1600 m, c 1700 m, d 1800 m, e 1900 m, and f 2000 m. The correlograms indicate two major changes across scenarios: (1) substantial decrease in the size (in cost distance) of the genetic neighborhood of positive genetic correlation and (2) increasing variability in genetic correlations at all lag distances
Discussion
Climate change and population connectivity in American marten
Recent research has shown that connectivity of American marten populations in the northern Rocky Mountains is driven by elevational gradients of landscape resistance (Wasserman et al. 2010). This relationship between gene flow and elevation probably reflects the species’ requirement for deep snowpacks, which are absent below roughly 1400 m in our study area. In addition, Wasserman (2008) found that marten select home ranges in unfragmented, mesic, middle elevation forest. Climate change is likely to both reduce snowpack depth and duration over relatively short time horizons, and lead to upward migration of optimal vegetation conditions over longer time scales. In the current landscape, resistance to gene flow is minimum at 1500 m and increases as a Gaussian function as elevations increase or decrease from that optimum (Wasserman et al. 2010).
In our simulations, we varied the optimal elevation across a range from 1500 to 2000 m, by 100 m steps. If we assume that marten are responding to snowpack and vegetation conditions that are driven by average temperature, these scenarios reflect temperature changes of approximately +0.65°C per 100 m scenario step and +3.25°C warming at the 2000 m optimum. The expected average temperature increase in this portion of the Rocky Mountains between 2000 and 2080 is +3.3°C (Littell et al. 2011). Therefore, our scenarios reflect realistic and rather modest range of potential effects of climate change on landscape resistance for marten. However, how rapidly this climate change affects marten connectivity will depend on whether connectivity is primarily driven by snowpack conditions or vegetation type distribution along elevation gradients. Snowpack will respond rapidly to climate change (given it is a yearly phenomenon) while vegetation will respond much more slowly, with time lags of up to several hundred years (Davis 1986; Baker 1995; Cushman et al. 2007).
By applying a least cost resistant kernel approach, we were able to map the portions of the landscape that would be predicted to be connected by dispersal at each scenario. The monotonic decrease in the area of predicted connected habitat from the current landscape suggests that climate driven changes in habitat connectivity may result in a 60% reduction in the area of the landscape occupied by connected marten habitat by 2080.
Fragmentation of marten habitat (as measured by number of isolated patches) increases nonlinearly, with a threshold appearing at scenario +300 m. Between the current and +300 m scenarios the number of isolated patches of connected habitat remains relatively constant at between 18 and 22. This suggests that expected warming between the current conditions and those expected in approximately 2040 will result in relatively little fragmentation of marten populations in the study area. In contrast, the number of isolated patches of habitat increases by over 100% (from 22 to 45) between scenarios +300 and +500 m. This suggests that extensive population fragmentation may be expected as a result of climate change between 2040 and 2080. It is interesting that while the area of marten habitat may decline smoothly with climate change, the fragmentation of the population is likely to change abruptly as average temperatures increase 1.5–2°C (Fig. 3).
Genetic consequences of population fragmentation resulting from climate change
The linear decline of allelic richness with decreasing habitat area suggests that as habitat is lost to climate change the total genetic diversity (as measured by number of alleles in the population) will decline roughly proportionally. The pattern of decline suggests that allelic richness is driven by habitat amount rather than fragmentation and no non-linear thresholds are expected where allelic diversity will drop dramatically with a small change in habitat amount or connectivity.
A substantially different pattern was seen in our simulations of expected heterozygosity. In contrast to allelic richness, expected heterozygosity showed a non-linear pattern of decrease across the climate change scenarios. This suggests that expected heterozygosity of the population may be more sensitive to habitat fragmentation, which also showed a threshold response beginning at the +300 m scenario, than to habitat loss per se.
The rapid non-linear decline of expected heterozygosity in scenarios +300 to +500 m and the monotonic decline in allelic diversity have major potential conservation implications. Loss of heterozygosity may result in inbreeding depression (Keller and Waller 2001). Inbreeding depression is the individual loss of fitness arising from deleterious recessive alleles or overdominance, and has been strongly linked to extinction risk in wild populations (O’Grady et al. 2006). In addition, the loss of allelic diversity predicted to occur in future landscape scenarios would be expected to reduce evolutionary potential and favor increased risk that deleterious alleles would become fixed in the population (Lacy 1997; Willi et al. 2006). The reduction in fitness caused by inbreeding depression may cause a cycle of ever declining population size and genetic diversity, ultimately resulting in population extirpation (Lacy 1997; Tanaka 2000).
In spatially structured populations experiencing habitat loss and fragmentation global measures of genetic change across the full population may not be sufficiently informative to evaluate conservation implications. In highly fragmented populations, the local genetic structure may be highly variable as functions of local landscape structure interacting with local population size and dispersal. Exploring the patterns of local genetic variation can help identify biological consequences of changes in the extent and pattern of habitat resulting from climate change (e.g. Shirk and Cushman 2011). The pattern in change in allelic richness indicates how reductions in habitat area and increases in habitat subdivision may affect genetic diversity in each scenario (Fig. 6). The change from the current scenario to +100 m results in substantial loss of allelic richness in small peninsular and island populations in the Selkirk Mountains, and several places in the Purcell mountains (green and blue areas in Fig. 6b). An area of the central Selkirk Mountains experiences moderate loss of allelic richness, indicating that the connection between the northern and southern Selkirk range is being reduced (green in Fig. 6b). This is interesting because it shows that even modest increase in average temperature (e.g. 0.65°C) can have relatively large effects on local population connectivity and resulting genetic structure. The patterns of loss across scenarios +100 to +500 m show progressive fragmentation where additional portions of the population become peninsular or insular and experience substantial reduction in allelic richness (Fig. 6c–f).
Decreasing genetic neighborhood size under climate change
In addition to reducing global allelic richness and expected heterozygosity, the simulated climate change scenarios also dramatically affected the size of local genetic neighborhoods. We defined local genetic neighborhoods as the area surrounding an individual in which a positive genetic correlation was expected. In the current landscape there is a very long range of genetic connectivity in the current landscape, indicating likely extensive genetic exchange within ranges (e.g. within the Selkirk Mountains or within the Cabinet Mountains), but lower genetic exchange between the ranges. This is consistent with the empirical data from the study area, which indicate a range of genetic correlation of approximately 83,000 cost units (Wasserman 2008), producing moderate genetic differentiation across the study area with the largest genetic differences seen between the Selkirk and Purcell Mountains (Wasserman et al. 2010).
The range of genetic autocorrelation declined dramatically across climate change scenarios. By the +500 m scenario, corresponding to expected climate in year 2080 (+3.3°C, Littell et al. 2011), there was an approximately 46% reduction in genetic neighborhood size, reflecting genetic isolation resulting from habitat subdivision. As climate change progresses in our simulations, the marten population becomes progressively fragmented, particularly after scenario +300 m (year 2040). This results in genetic isolation of small groups of martens on the landscape, in turn reducing the extent of genetic neighborhoods. This increases the strength of genetic correlation between martens at small distance classes, by reducing long range dispersal, and increases variation in strength of genetic autocorrelation at all distance classes. The large reduction in genetic neighborhood size is closely tied to the observed decreases in allelic richness and expected heterozygosity. Together these indicate that climate change is likely to fragment the marten population in the study area, reduce the size of genetically connected neighborhoods, and result in substantial loss of genetic diversity and heterozygosity.
Scope and limitations
Uncertainty in the relationship between expected warming and change in landscape resistance
We utilize a simple method of projecting future landscape resistance in this study, which is enabled by the relatively simple relationship between current landscape resistance and genetic structure of the species. Landscape resistance for American marten in this study area is univariate function of elevation (Wasserman et al. 2010). There is some uncertainty in the temperature lapse rate as elevation increases in the study area. Based on standard environmental lapse rate of −6.5°C/km increase in elevation (e.g., Willmott and Matsuura 1995), each +100 m step in our analysis represents an increase of 0.65°C. However, average lapse rates in the maritime Pacific Northwest are often shallower (Minder et al. 2010). Under this calculation each 100 m increment of our simulation would equate to a 0.45°C increase, with +500 m equating to a 2.25°C total warming. If lapse rates are this flat, our simulation of fragmentation is excessively conservative, and the expected impacts of the forecast +3.3°C warming by 2080 are likely to be greater than reported here.
There is also substantial variability in lapse rates seasonally. We do not know if the observed relationship between elevation and landscape resistance is associated more strongly with winter or summer climate. We suspect it is likely more associated with winter, given the strong linkage between snowpack and marten occurrence and between snowpack and mesic forest vegetation. Across the year, spring lapse rates are steepest (Minder et al. 2010 found 6.5–7.5°C/km average), while summer rates are flattest. If marten are most sensitive to winter temperatures (related to snowpack), the expected lapse rate may be flatter than considered in this study. This would result in our study underestimating the effects of climate change on habitat loss and fragmentation of this species, as a flatter lapse rate would result in a larger upward shift of optimum dispersal habitat per unit of warming.
Elevation is closely associated with snowpack and gradients of forest vegetation composition and structure. Deep persistent snow pack is a critical habitat element for the species. It excludes predators (e.g. Coyote, Canis latrans), and provides high-quality hunting conditions via subnivean space (Buskirk and Ruggiero 1994). In our study area Subalpine fir (Abies lasiocarpa) and Engelmann spruce (Picea engelmannii) are codominant above 1500 m, and a diverse mixed forest of Western Larch (Larix occidentalis), Western Red cedar (Thuja plicata), and other conifer dominates between 1300 and 1500 m (Evans and Cushman 2009). These forest types have been found to be strong predictors of occurrence of marten in this study area (Wasserman 2008). Thus the observed relationship between elevation and gene flow probably reflects a combination of the effects of snowpack and vegetation gradients on dispersal. Assuming the ecological processes that control landscape resistance change monotonically with changing climate, it is reasonable to project landscape resistance into the future by linking the landscape resistance function to current temperature and moving it upwards in elevation to reflect different degrees of warming. How rapidly this climate change affects marten connectivity will depend on whether connectivity is primarily driven by snowpack conditions or vegetation type distribution along elevation gradients. Snowpack will respond rapidly to climate change (given it is a yearly phenomenon) while vegetation will respond much more slowly, with time lags of up to several hundred years (Davis 1986; Baker 1995; Cushman et al. 2007).
Conclusions
This study is the first to combine empirically derived estimates of landscape resistance, formal projection of these resistance surfaces under a realistic range of climate change scenarios, simulation of habitat connectivity, simulation of population-level and local genetic diversity and evaluation of changes in the extent of genetic neighborhoods in the current landscape and each future scenario. The combination of these enables this study to quantitatively project effects of climate change on habitat area and connectivity for a climate sensitive species, and evaluate the implications of these changes for genetic diversity. Our results indicate that American marten in the northern Rocky Mountains of the United States are likely to be highly sensitive to climate change. We predict large reductions in the area and connectivity of dispersal habitat under a plausible range of climate change scenarios. Expected climate change by year 2080 may reduce the extent of dispersal habitat in our study area by over 50% and more than double the number of isolated habitat patches. These changes in habitat extent and pattern are likely to dramatically reduce allelic richness and expected heterozygosity. These genetic changes may have severe consequences. Inbreeding depression has been strongly linked to extinction risk and the loss of allelic diversity reduces evolutionary potential and the risk fixation of deleterious alleles. The reduction in fitness due to inbreeding depression and loss of allelic diversity may promote a cycle of ever declining population size and genetic diversity, ultimately resulting in population extirpation in what has been termed an extinction vortex.
References
Baker WL (1995) Longterm response of disturbance landscapes to human intervention and global change. Landscape Ecol 10:143–159
Broquet T, Johnson CA, Petit E, Thompson I, Burel F, Fryxell JM (2006) Dispersal and genetic structure in the American marten, Martes americana. Mol Ecol 15:1689–1697
Bruggeman DJ, Wiegand T, Fernandez N (2010) The relative effects of habitat loss and fragmentation on population genetic variation in the red-cockaded woodpecker (Picoides borealis). Mol Ecol 19:3679–3691
Bull EL, Heater W (2001) Home range and dispersal of the American marten in northeastern Oregon. Northwest Nat 82:7–11
Buskirk SW, Ruggiero LF (1994) The American marten. In: Ruggiero LF, Aubry KB, Buskirk SW, Lyon LJ, Zielinski WJ (eds) American marten, fisher, lynx, and wolverine in the western United States. General Technical Report, RM-254. U.S. Department of Agriculture, Forest Service, Rocky Mountain Forest and Range Experiment Station, Fort Collins, CO
Compton B, McGarigal K, Cushman SA, Gamble L (2007) A resistant kernel model of connectivity for vernal pool breeding amphibians. Conserv Biol 21:788–799
Cushman SA, Landguth EL (2010) Spurious correlations and inference in landscape genetics. Mol Ecol 19:3592–3602
Cushman SA, Schwartz MK, Hayden J, McKelvey KS (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486–499
Cushman SA, McKenzie D, Peterson DL, Littell JS, McKelvey KS (2007) Research agenda for integrated landscape modelling. USDA For. Serv. General Technical Report RMRS-GTR-194
Cushman SA, Chase MJ, Griffin C (2010a) Mapping landscape resistance to identify corridors and barriers for elephant movement in southern Africa. In: Cushman SA, Huettman F (eds) Spatial complexity, informatics and wildlife conservation. Springer, Tokyo, pp 349–368
Cushman SA, McGarigal K, Gutzwiller K, Evans J (2010b) The gradient paradigm: a conceptual and analytical framework for landscape ecology. In: Cushman SA, Huettman F (eds) Spatial complexity, informatics and wildlife conservation. Springer, Tokyo, pp 83–110
Cushman SA, Landguth EL, Flather CH (2011a) Climate change and connectivity: assessing landscape and species vulnerability. Final Report to USFWS Great Plains Landscape Conservation Cooperative
Cushman SA, Wasserman TN, McGarigal K (2011b) Landscape fire and wildlife habitat, pp 223–254. In: McKenzie D, Kellog L-K (eds) The Landscape Ecology of Fire. Ecology Studies 213, Springer
Davis M (1986) Climatic instability, time tags, and community disequilibrium. In: Diamond J, Case T (eds) Community ecology. Harper and Row, New York, pp 269–284
de Vos A (1951) Overflow and dispersal of marten and fisher from wildlife refuges. J Wildl Manag 15:164–175
Elsner MM, Cuo L, Voisin N, Deems J, Hamlet AF, Vano JA, Mickelson KEB, Lee SY, Lettenmaier DP (2010) Implications of 21st century climate change for the hydrology of Washington State. Clim Change 102(1–2):225–260. doi:10.1007/s10584-010-9855-0
Epperson BK, McRae B, Scribner K, Cushman SA, Rosenberg MS, Fortin M-J, James PMA, Murphy M, Manel S, Legendre P, Dale MRT (2010) Utility of computer simulations in landscape genetics. Mol Ecol 19:3540–3564
ESRI (2005) ARCGIS. Environmental systems research incorporated. Redlands, CA
Evans JS, Cushman SA (2009) Gradient modeling of conifer species using random forests. Landscape Ecol 24:673–683
Gardner RH, Gustafson EJ (2004) Simulating dispersal of reintroduced species within heterogeneous landscapes. Ecol Model 171:339–358
IPCC (2007) In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, Miller HL (eds) Climate change 2007: the physical science basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge
Iverson LR, Prasad A, Schwartz MW (1999) Modeling potential future individual tree-species distributions in the eastern United States under a climate change scenario: a case study with Pinus virginiana. Ecol Model 115:77–93
Keller L, Waller D (2001) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Lacy RC (1997) Importance of genetic variation to the viability of mammalian populations. J Mammal 78:320–335
Landguth EL, Cushman SA (2010) CDPOP: an individual-based, cost-distance spatial population genetics model. Mol Ecol Resour 10:156–161
Landguth EL, Cushman SA, Murphy MA, Luikart G (2010a) Relationships between migration rates and landscape resistance assessed using individual-based simulations. Mol Ecol Resour 10:854–862
Landguth EL, Cushman SA, Luikart G, Murphy MA, Schwartz MK, McKelvey KS (2010b) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191
Littell JS, Elsner MM, Mauger G, Lutz E, Hamlet AF, Salathé E (2011) Regional climate and hydrologic change in the northern US Rockies and Pacific Northwest: internally consistent projections of future climate for resource management. http://cses.washington.edu/picea/USFS/pub/Littell_etal_2010/Littell_etal._2011_Regional_Climatic_And_Hydrologic_Change_USFS_USFWS_JVA_17Apr11.pdf
McGarigal K, Cushman SA, Neel MC, Ene E (2002) FRAGSTATS: spatial pattern analysis program for categorical maps. Computer software program produced by the authors at the University of Massachusetts, Amherst. Available at www.umass.edu/landeco/research/fragstats/fragstats.html
McKelvey KS, Copeland JP, Schwartz MK, Littell JS, Aubry KB, Squires JR, Parks SA, Elsner MM, Mauger GS (in press) Climate change predicted to shift wolverine distributions, connectivity, and dispersal corridors. Ecol Appl. doi:10.1890/10-2206.1
McKenzie D, Peterson DW, Peterson DL, Thornton PE (2003) Climatic and biophysical controls on conifer species distributions in mountain forests of Washington State, USA. J Biogeogr 30:1093–1108
Minder JR, Mote PW, Lundquist JD (2010) Surface temperature lapse rates over complex terrain: lessons from the Cascade Mountains. J Geophys Res 115:D14122. doi:10.1029/2009JD013493
Mote PW, Salathé EP (2010) Future climate in the Pacific Northwest. Clim Change 102(1–2):29–50. doi:10.1007/s10584-010-9848-z
O’Grady JJ, Brook BW, Reed DH, Ballou JD, Tonkyn DW, Frankham R (2006) Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol Conserv 133:42–51
Rehfeldt GE, Crookston NL, Warwell MV, Evans JS (2006) Empirical analysis of plant-climate relationships for the western United States. Int J Plant Sci 167:1123–1150
Shirk AJ, Cushman SA (2011) sGD software for estimating spatially explicit indices of genetic diversity. Mol Ecol Resour 11:923–934
Tanaka Y (2000) Extinction of populations by inbreeding depression under stochastic environments. Popul Ecol 42:55–62
Tomson SD (1999) Ecology and summer/fall habitat selection of American marten in Northern Idaho. MS thesis, University of Montana, Missoula
Wasserman TN (2008) Habitat relationships and gene flow of Martes americana in northern Idaho. MS thesis. Western Washington University, Bellingham
Wasserman TN, Cushman SA, Wallin DO (2010) Spatial scaling and multi-model inference in landscape genetics: Martes americana in northern Idaho. Landscape Ecol 25:1601–1612
Wiens JA, Stralberg D, Jongsomjit D, Howell C, Snyder MA (2009) Niches, models and climate change: assessing the assumptions and uncertainties. Proc Natl Acad Sci USA 106:19729–19736
Willi Y, Van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37:433–458
Willmott CJ, Matsuura K (1995) Smart interpolation of annually averaged air temperature in the United States. J Appl Meteorol 34:2577–2586
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wasserman, T.N., Cushman, S.A., Shirk, A.S. et al. Simulating the effects of climate change on population connectivity of American marten (Martes americana) in the northern Rocky Mountains, USA. Landscape Ecol 27, 211–225 (2012). https://doi.org/10.1007/s10980-011-9653-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-011-9653-8