Abstract
Fine-scale landscape change can alter dispersal patterns of animals, thus influencing connectivity or gene flow within a population. Furthermore, dispersal patterns of different species may be influenced by the landscape in varying ways. Our research first aimed to examine whether the spatial genetic structure within populations of closely related bird species differs in response to the same landscape. Second, we examined whether individual-level movement characteristics are a mechanistic driver of these differences. We generated a priori predictions of how landscape features will influence dispersal (particularly the response of individuals to habitat boundaries both natural and human-induced) based on a movement model developed by Fahrig (Funct Ecol 21:1003–1015, 2007). This model allowed us to predict genetic relatedness patterns in populations of two passerine bird species with different life-history traits from Queensland, Australia (yellow-throated scrubwren Sericornis citreogularis, a habitat specialist; white-browed scrubwren Sericornis frontalis, a habitat generalist). We quantified our predictions using cost-distance modelling and compared these to observed pairwise genetic distances (a r ) between individuals as calculated from microsatellite markers. Mantel tests showed that our a priori models correlated with genetic distance. Euclidean distance was most closely correlated to genetic distance for the generalist species (r = 0.093, P = 0.002), and landscape models that included the avoidance of unsuitable habitat were best for the specialist species (r = 0.107, P = 0.001). Our study showed that predictable movement characteristics may be the mechanism driving differences in genetic relatedness patterns within populations of different bird species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An urban growth characteristic of many metropolitan areas is extension into new undeveloped sites, particularly in Australia and the United States (Bunker and Houston 2003; Low Choy 2006). This development commonly occurs in otherwise intact habitat, and can result in housing and amenities scattered throughout natural areas (McKenzie 1996; Houston 2005). The small scale landscape change and habitat fragmentation that results from such development has negative implications for the conservation of birds and other animals (Cushman 2006; Kath et al. 2009). Habitat loss can decrease the size of a population, whereas habitat fragmentation alters dispersal patterns severing populations into smaller less viable parts (Wilcox and Murphy 1985; Bunker and Houston 2003; Kath et al. 2009). Furthermore, landscape heterogeneity and fragmentation can affect both the demography and genetic patterns of populations (e.g. Cushman et al. 2006; Pérez-Espona et al. 2008; Bruggeman et al. 2010). We are, however, still developing an understanding about how small scale landscape patterns influence a population’s genetic/spatial structure (landscape pattern, in this paper, refers to patterns of vegetation types, topography, clearing etc.). Information on how different species respond to landscape pattern (and functional connectivity) at this scale will provide a basis for planning guidelines to prevent (or repair) early effects of fragmentation (Holderegger and Wagner 2008).
Animal tracking studies suggest that at a small scale, landscape pattern influences different species in different ways; for example, the response of individuals to habitat/non-habitat boundaries differs among species (With 1994; Fahrig 2007; Lindenmayer and Fisher 2007; Gillies and Clair 2008; Bruggeman et al. 2010). This variability among species presents difficulties for planning rehabilitation efforts or landscape development. In a seminal paper, however, Fahrig (2007) suggested that differences in individual level movement characteristics may follow general rules according to a grouping system based on species life-history traits. Fahrig’s paper synthesised evidence from animal movement studies, and presented a general model which describes how characteristics of a species’ ‘natural landscape’ influence the evolution of movement parameters and dispersal behaviour of individuals. The term ‘natural landscape’ refers to whether a species’ preferred habitat types are continuous, patchy or ephemeral in the landscape where they evolved.
In general, Fahrig’s model predicts that a species which naturally inhabits a highly patchy landscape will have greater avoidance of habitat boundaries, whereas a species which naturally occurs in areas of more continuous habitat will have a less severe boundary response. Fahrig proposes that this differential movement response of species is linked to the level of risk an individual experiences outside of the preferred habitat type (i.e. in the ‘matrix’). Supporting this idea, fragmentation studies have cited movement characteristics as one of the possible causes of a species-specific susceptibility to habitat fragmentation (Andrén et al. 1997; Sekercioglu et al. 2002). Fahrig’s model presents an opportunity to explore individual level movement characteristics, particularly a species’ response to non-habitat boundaries, as a potential mechanistic driver of observable spatial structure of populations, and also the susceptibility of species to fine-scale habitat fragmentation.
Developments in individual-based genetic techniques (measurements of relatedness between pairs of individuals of a species) have created significant opportunities for investigating dispersal routes of animals (Streiff et al. 1998; Rousset 2000; Vos et al. 2001; Broquet et al. 2006; Cushman et al. 2006). Gene flow is understood to occur step-by-step as individuals disperse across subsequent generations, and thus the greater the number of steps between two individuals in a particular time frame the greater the genetic distance will be between them (Wright 1943; Coulon et al. 2004). When coupled with landscape information, individual based genetic differences provide a means to examine hypotheses of dispersal routes (Coulon et al. 2004; Cushman et al. 2006, 2008). The empirical studies that have utilised these techniques confirm that individual-based genetic relatedness is a reasonable proxy for dispersal routes (Coulon et al. 2004; Broquet et al. 2006), as it has been found to correlate well with vegetation patterns within the landscape. But, the way in which different species respond to the same landscape, and the mechanism driving these differences, remain to be examined.
In this study we had two main objectives. The first was to examine whether the spatial genetic structure within populations of two closely related passerine bird species (both within the genus Sericornis) differed in response to the same landscape, and further, whether there were sex differences in these responses. Each species studied had a different ‘natural landscape’. Our second objective was to examine whether individual level movement characteristics were a potential mechanistic driver of genetic differences. To address this problem, we used Fahrig’s model to make specific a priori predictions about how landscape features such as habitat boundaries (both natural and human-induced) likely affected dispersal routes and, thus, genetic relatedness patterns of the two bird species of interest. This approach, where specific falsifiable hypotheses are generated, is considered a relevant approach for landscape genetic studies to avoid spurious results (Segelbacher et al. 2010). We used individual-based genetic data collected from a single population of each species in the same landscape to confirm the validity of our model predictions. This study may provide a means to generalise the response of bird/animal populations to landscape pattern at small spatial scales.
Materials and methods
Study area, species and development of an a priori model
Our study area was located within the D’Aguilar National Park and the adjoining Brisbane Forest Park, north of Brisbane city in South East Queensland, Australia (Fig. 1). The primary vegetation types are eucalypt woodland and rainforest/wet sclerophyll forest, and there are natural abrupt boundaries between these vegetation types. Within the large natural forest are small sections that have experienced high levels of human disturbance; there is a main road through the centre of the park and many cleared or housed areas have existed for up to 150 years. Associated with these human altered areas are vegetation/clearing boundaries.
We selected two bird species for this study. The first is the white-browed scrubwren (Sericornis frontalis), a generalist species that utilises many vegetation types providing there is some shrubby understory present (Higgins and Peter 2002). The matrix in the study region for the white-browed scrubwren can be considered high quality, as it often contains small scattered shrubby areas that the species can be found foraging in (Fig. 2; Table 1). The second species is the yellow-throated scrubwren (Sericornis citreogularis), a specialist species restricted to dense rainforest (the surrounding ‘matrix’ can therefore be considered low quality), and whose natural habitat is stable but patchy (Fig. 2; Table 1) (Higgins and Peter 2002). The two species are closely related, representing two of the three species in the genus Sericornis, and are of similar body size (Higgins and Peter 2002). Due to differences in feeding preferences (Higgins and Peter 2002) they are unlikely to be direct competitors.
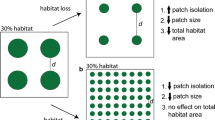
Our model describing animal movement as a function of species’ landscapes (derived from Fahrig 2007). The diagram shows our extension of Fahrig’s principles to make predictions on how a species’ movement characteristics and local vegetation patterns influence within-population genetic patterns of a specialist and generalist species. Notably, we predict that the strength of a species’ boundary response will define whether realised dispersal tends to occur through unsuitable habitat or not
Based on the life-history characteristics of the white-browed and yellow-throated scrubwren, we used Fahrig’s model (2007) to generate a priori expectations of how landscape pattern would affect individual relatedness patterns of these two species (Fig. 2). Our model was built on the basic assumption that movement characteristics of individuals during dispersal will determine patterns of relatedness within a population through the step-by-step process of dispersal (Wright 1943; Coulon et al. 2004). For instance, if individuals of a species are strongly repelled by inhospitable ‘matrix’ (i.e. they have a strong boundary response to unsuitable habitat), then dispersing individuals will also be more likely to preferentially travel through and settle in suitable habitat. Individuals are therefore likely to be more closely related along threads of suitable habitat rather than across even small amounts of unsuitable habitat.
Creating cost-surfaces to quantify model predictions
To quantify the a priori predictions and apply them to the species of interest, we used least-cost modelling. This modelling approach allows the user to determine the minimum distance between two points while taking into account areas which may be more costly to cross or are simply more resistant to crossing (Adriaensen et al. 2003; Chardon et al. 2003). This approach involves first creating a cost surface which is a simplified grid-based interpretation of the landscape. Each grid cell is assigned a ‘cost value’. An algorithm is then used to determine the least cost distance between two points.
We first created a cost surface for both species based on the a priori model described in Fig. 2. For the white-browed scrubwren (the generalist species), we predicted that Euclidean distance should provide the best correlation with genetic relatedness between individuals. As such, the a priori cost surface for the white-browed scrubwren was a uniform surface where all landscape features were considered equally permeable to dispersal and gene flow, and all grid cells were assigned the same cost value (see Model 1 of the alternative model set presented in Table 2 for cost values). For the yellow-throated scrubwren, we expected that patterns of suitable habitat would determine dispersal patterns, where the most suitable habitat (densely vegetated drainage routes in the area of interest) provided the best facilitator of dispersal and gene flow. The a priori cost surface for this species was therefore characterised by high resistance of unsuitable habitat to movement, a low level of resistance by suitable habitat, and even lower resistance along drainage lines (see Model 4 from the alternative model set described in Table 2).
We created four additional alternative landscape models for each species to compare against our a priori landscape models. These additional models considered a range of landscape features, including unsuitable/suitable habitat, drainage lines, and human altered areas (which included roads, housing, and other cleared areas). We took a factorial approach to assigning cost values to the landscape features within these models, assigning a high and low value for each feature (Table 2). This approach allows the examination of the relative importance of each factor, rather than its exact level of resistance to dispersal (Cushman et al. 2006). This process resulted in 17 possible landscape models to be tested for each species (Table 2)—five principle landscape models with sub-models of varying cost values.
To generate the cost surfaces, we used vector data on vegetation, topography and human altered areas in ARCGIS 9.3 (ESRI). We first built a suitable habitat map for each species using species information from Higgins and Peter (2002). To do this, we developed a list of suitable habitat types and extracted relevant areas from a detailed vegetation map generated by local government (1:50,000; Regional Ecosystems version 5.2; Environmental Protection Agency 2007). The user’s accuracy (probability that the actual vegetation type is what the map says it is) of this broadly classified map was 95%. Topographical layers (1:250,000; Geoscience Australia 2006) provided drainage line locations. We generated our own ‘human altered areas’ vector layer by creating polygons representing housing, clearings and major roads with high resolution from Google Earth® imagery (Spot image, captured April 16, 2007). Due to expected errors in projection which can occur with Google Earth® imagery, the human altered areas layer was ground truthed and rectified using prominent human-made landmarks (recognisable houses and forestry road corners) across the studied landscape. All vector layers were converted to grid with a resolution of 10 m. This value is equal to the finest scale features within our input surfaces, potentially allowing us to detect the importance of biologically relevant, but small, landscape features (Adriaensen et al. 2003). These grid layers were combined, and features assigned values (as per Table 2) within ARCGIS 9.3 (ESRI) to create our final cost surfaces.
Field data collection and genetic analysis
We sampled individuals from a variable part of the study area on a grid of approximately 1 km between sampling points (Fig. 1). This scale was chosen as it is significantly larger than an individual’s territory size for both species, but less than an approximated dispersal distance as calculated according to Bowman (2003). We covered a landscape approximately 14 km in length and up to 3 km wide (Fig. 1). Trapping was carried out early in the single breeding season (spring to early summer 2007/2008), when most individuals were likely settled and not actively dispersing. At each location a male and female, where possible, of each species were live-trapped (mist-netted). Each individual was banded and measured to prevent double sampling. Approximately 10–20 μl of blood was taken from the brachial vein (according to the technique described by Arctander 1988) and stored in Queens Lysis Buffer (Seutin et al. 1991). A total of 41 adult white browed scrubwren (19 female, 22 male) and 41 adult yellow-throated scrubwren (15 female, 26 male) were sampled at 21 locations. A salting out method was used to extract DNA from these samples (Nicholls et al. 2000). Twelve microsatellite markers were screened for amplification for both species (markers previously isolated for the large-billed scrubwren, Sericornis magnirostris; Bardeleben et al. 2005). Seven microsatellite markers for the white-browed scrubwren and six for the yellow-throated scrubwren amplified and were polymorphic. Polymerase Chain Reaction (PCR) conditions followed Bardeleben et al. (2005) using BioTaq DNA polymerase (Bioline). Fragments were visualised using a Corbett GS2000 acrylamide gel system.
We examined whether the markers were in Hardy–Weinberg equilibrium with an exact test in GENEPOP version 4.0 (Raymond and Rousset 1995) and carried out sequential Bonferroni correction for multiple tests (Rice 1989). To further test the microsatellite data, we looked for evidence of admixture (i.e. multiple populations), checked whether close relatives were sampled and also tested for null alleles in the data set. Details on these analyses are presented in the supplementary material.
We chose to use a r (Rousset 2000) as the measure of genetic distance between individuals. This metric is known to provide a reliable estimate of genetic difference between individuals and has been repeatedly used in individual based genetic studies (Coulon et al. 2004; Broquet et al. 2006). We calculated a r between individuals using the method described by Rousset (2000) in GENEPOP 4.0. This provided a matrix of pairwise genetic differences between all pairs of individuals within each species. Other distance indices such as Nei’s D (Hendrik 1983) would have produced nearly identical results as the Pearson correlation coefficient between a r and Nei’s D in this study was more than 0.98 for both species.
Model assessment
The geographic least-cost distances between individuals for both species were calculated for all 17 landscape models using PATCHMATRIX (Ray 2004), an extension to ARCVIEW 3.3 (ESRI). All geographic distances were loge-transformed.
We examined the relationship between a r and the geographic distances calculated from each landscape model using Mantel tests (Mantel 1967) in GENALEX version 6.0 (Peakall and Smouse 2006; permutations = 9999). These tests use a modified regression analysis between two data sets to assess autocorrelation between two matrices; the rows and columns of the matrices are randomly re-sampled to calculate r (the correlation coefficient) and the Monte Carlo procedure estimates significance of the test. We carried out Mantel tests for all samples of each species as well as each species separated by sex. We used the Bonferroni correction to adjust P values for multiple tests (α = 0.05). We ranked each of the models from 1 to 17 based on Mantel correlation coefficients, and then by the P value of the test (a higher coefficient and lower P value will result in a higher ranking). This allowed us to examine which sub-models had the greatest correlation with the genetic data. In this least-cost distance modelling approach, all features in each landscape model are combined to produce a single resistance hypothesis. As such there is no inflation of explained variance due to the number of factors involved (Cushman 2006).
Partial Mantel tests allow the user to test for a relationship between two variables, while controlling for a third. We used partial Mantel tests within a causal modelling framework, where the correlation between each landscape model and a r was tested with each of the other models partialled out. The results from this analysis can be used to assess the degree of support for each model (Legendre and Troussellier 1988; Cushman et al. 2006; Lada et al. 2008). This is because a positive and significant partial Mantel correlation coefficient indicates that the landscape model tested provides a significantly better fit to the genetic data than the model that is partialled out. Within this causal modelling framework we used only the highest ranking sub-model from each of the five landscape model sets. To evaluate support for each model we assessed the correlation coefficients and P values. All partial Mantel tests were carried out in the package ‘vegan’ (Oksanen et al. 2009) within R package version 2.2 (R Development Core Team 2005).
Results
Genetic analyses
All microsatellite loci had between two and 33 alleles. Observed heterozygosity (Ho) was significantly lower than expected heterozygosity (He) in one locus for the yellow-throated scrubwren (AACC-33) and two for the white-browed scrubwren (AACC-33, AACC-1). When these loci were removed from the analysis there was no difference in top ranking models (these results are presented in the supplementary material). There was no evidence that parent/offspring or siblings were sampled (see the supplementary material), and no null alleles were detected. The results indicated that individuals were sampled from a single population for both species.
Overall model test
The highest ranking landscape model was Model 1 (Euclidean distance) for white-browed scrubwren (the habitat generalist, r = 0.093, P = 0.002), and Model 4a (the more complex model with predicted resistance to natural habitat boundaries) for the yellow-throated scrubwren (the habitat specialist, r = 0.107, P = 0.001). No other landscape models were significant for the white-browed scrubwren, but for the yellow-throated scrubwren Models 3b and 5a were also significant (Tables 3, 4).
We observed sex based differences in the relationship between geographic distance and genetic distance in both species. No significant correlation with geographic distance or any other model was found for white-browed scruwbren males and yellow-throated scrubwren females. However, there were significant results for white-browed scruwbren females and yellow-throated scrubwren males with the highest ranking models Model 1 (Euclidean distance) and Model 4a (resistance to natural habitat boundaries) respectively (Table 2).
The partial Mantel tests showed that for the yellow-throated scrubwren, both Model 4a and Model 5a (where human altered areas were also considered) improved significantly on the relationship with a r over all other tested models, indicating that vegetation had an important effect on within population genetic structure in this species and that drainage lines provide corridors for gene-flow. We were unable to distinguish whether either Model 4a or Model 5a were better, as the partial Mantel test correlation coefficient between them was not significant. For the white-browed scrubwren, Euclidean distance (Model 1) was much better at explaining a r than the more complex Models 4c and 5e. Further, when the other models were tested against a r with Euclidean distance partialled out, no results were significant. There was thus no evidence that landscape complexity was more important than distance alone in determining the genetic structure of this species. In both species Model 2, which only included areas altered by humans, did not have a significant relationship with genetic distance.
Discussion
Information on the way that landscape patterns influence within-population genetic structure of different species could provide a key to understand the effect of early stage habitat fragmentation on processes such as dispersal (Cushman and McGarigal 2002; Proctor et al. 2005; Bruggeman et al. 2010). In this paper we have shown that the relationship between landscape features and genetic patterns within populations differ even between two closely related species in the same landscape. Furthermore, we have presented evidence that differential movement characteristics may be the mechanism driving this difference. This result highlights the importance of considering differences in species life-history traits when de-fragmenting landscapes.
In support of our a priori predictions, we found that Euclidean distance best described genetic patterns for the white-browed scrubwren, a species that naturally occurs in patchy habitat with high quality, low-risk matrix (and hence was predicted to have a weak boundary response). The more complex model that included patterns of suitable habitat provided the best correlation for the yellow-throated scrubwren, a species with a patchy natural landscape, high-risk matrix and predicted strong avoidance response to natural habitat boundaries. These results provide some clues as to how each species might respond to landscape change. For example, if a species has a strong avoidance response to natural boundaries and this impacts on within-population spatial genetic structure, the creation of new boundaries should hinder normal dispersal patterns in the long-term, fragmenting the population into smaller and correspondingly more ‘at risk’ populations (Wilcove et al. 1986; Fahrig 2003). However, a species such as the white-browed scrubwren with a weak boundary response to natural habitat boundaries may be more resilient to fragmentation as dispersal processes could be maintained. Our results also support other research that suggested habitat specialists may be more at-risk from habitat fragmentation. For instance, Vergara and Armesto (2009) found that specialist species are more affected by landscape pattern across spatial scales, and other studies found that habitat specialists are less likely to cross unsuitable habitat areas to recolonise habitat remnants (Villard and Taylor 1994; Joshi et al. 2006). To ensure long term persistence of specialist species such as the yellow-throated scrubwren, direct connections among habitat patches should be the best way to ensure population maintenance.
We also assessed whether landscape models which incorporated human altered areas provided predictive accuracy (Model 2). Our results did not show any significant influence of this landscape feature in both species. This was surprising, as our a priori predictions expected that landscape change would have an early and significant influence on the yellow-throated scrubwren with its strong boundary response. If this study was replicated in a region where human induced landscape change was larger or occurred earlier (i.e. more than 100–150 years ago), a contrasting result may emerge.
We did observe a difference in the relationship between geographic distance and genetic distance for males and females of both the yellow-throated scrubwren and white-browed scurbwren, which may indicate that each species has sex-biased dispersal, although we could not explore this pattern in detail due to small sample sizes.
Genetic studies at the individual-based level such as this one present two main challenges which need to be considered when interpreting results. First, as this study presents, the correlation coefficients from Mantel/partial Mantel tests were low in the comparisons between genetic distance (a r ) and geographic distances. This is a characteristic of many studies using individual-based genetics techniques, probably because genetic drift and sampling error at this level result in high variance in spatial genetic structure. For example, Coulon et al. (2004) achieved correlation coefficients of 0.0001–0.031 for Mantel tests between individual-based genetic and geographic distances for roe deer, and in a study on the American marten the coefficients were between 0.0002 and 0.0043 (Coulon et al. 2004; Broquet et al. 2006). However, their results are generally considered biologically meaningful providing the relationship is significant as it depends directly on dispersal patterns (Broquet et al. 2006). One way to improve the strength of results in such studies, may be to utilise a greater number of microsatellite loci (Koskinen et al. 2004). However, in our study six and seven loci were sufficient to provide significant results. The second challenge presented is that populations sampled at a small spatial scale can result in relatively small sample sizes. A greater number of individuals sampled might strengthen our results, though the number of sampling units in our study is still larger than in many population-level cost-distance studies—these studies also tend to use relatively few sample units and yet provide insights into landscape level processes. Individual-based simulation approaches (e.g. Landguth and Cushman 2010), will provide a means to further explore the relationship between landscape and genetic patterns within populations.
Conclusions
In this study we demonstrated that two co-occurring bird species are affected in different ways by the same landscape. Furthermore, we found support for the hypothesis that individual-level movement-response to landscape features is a likely mechanism driving these differences. Importantly, our model also provides a context for studies where no relationship between landscape pattern and genetic structure is found, as this was an a priori expected outcome for one of the species in this study.
Our study has implications for conservation management, particularly for developing ways to prevent or repair habitat fragmentation for different species. For example, at the within-population scale, vegetation corridors containing suitable habitat could be the best way to maintain dispersal patterns for species that naturally occur in patchy habitat and consequently have a strong boundary avoidance response. However, this may not be necessary for generalist species with a weak boundary response. The predictions presented in this study using Fahrig’s model (2007) could be accommodated to other species and differing landscapes.
References
Adriaensen F, Chardon JP, De Blust G, Swinnen E, Villalba S, Gulinck H, Matthysen E (2003) The application of ‘least-cost’ modelling as a functional landscape model. Landsc Urban Plan 64:233–247
Andrén H, Delin A, Seiler A (1997) Population response to landscape changes depends on specialization to different landscape elements. OIKOS 80:193–196
Arctander P (1988) Comparative studies of avian DNA by restriction fragment length polymorphism analysis: convenient procedures based on blood samples from live birds. J Ornithol 129:205–216
Bardeleben C, Gray MM, Austin J, de Rosario IA (2005) Isolation of polymorphic tetranucleotide microsatellite for the large-billed scrubwren (Sericomis magnirostris). Mol Ecol Notes 5:143–145
Bowman J (2003) Is dispersal distance of birds proportional to territory size? Can J Zool 81:195–202
Broquet T, Ray N, Petit E, Fryxell JM, Burel F (2006) Genetic isolation by distance and landscape connectivity in the American marten (Martes americana). Landscape Ecol 21:877–889
Bruggeman DJ, Wiegand T, Fernández N (2010) The relative effects of habitat loss and fragmentation on population genetic variation in the red-cockaded woodpecker (Picoides boralis). Mol Ecol 19:3679–3691
Bunker R, Houston P (2003) Prospects for the rural–urban fringe in Australia: observations from a brief history of the landscapes around Sydney and Adelaide. Aust Geogr Stud 41:303–323
Chardon JP, Adriaensen F, Matthysen E (2003) Incorporating landscape elements into a connectivity measure: a case study for the speckled wood butterfly (Pararge aegeria L.). Landscape Ecol 18:561–573
Coulon A, Cosson JF, Angibault JM, Cargnelutti B, Galan M, Morellet N, Petit E, Aulagnier S, Hewison AJM (2004) Landscape connectivity influences gene flow in a roe deer population inhabiting a fragmented landscape: an individual-based approach. Mol Ecol 13:2841–2850
Cushman SA (2006) Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol Conserv 128:231–240
Cushman SA, McGarigal K (2002) Hierarchical, multi-scale decomposition of species–environment relationships. Landscape Ecol 17:637–646
Cushman SA, McKelvey KS, Hayden J, Schwartz MK (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486–499
Cushman SA, McKelvey K, Schwartz MK (2008) Using empirically derived source-destination models to map regional conservation corridors. Conserv Biol 23:368–376
Environmental Protection Agency (2007) Regional Ecosystem Description Database (REDD). Version 5.2. Updated June 2007. Database maintained by Queensland Herbarium. Environmental Protection Agency, Brisbane
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Fahrig L (2007) Non-optimal animal movement in human-altered landscapes. Funct Ecol 21:1003–1015
Fischer J, Lindenmayer DJ (2007) Landscape modification and habitat fragmentation: a synthesis. Glob Ecol Biogeogr 16:265–280
Geoscience Australia (2006) GEODATA TOPO 250K Series 3. Geoscience Australia, Canberra
Gillies CS, Clair CCS (2008) Riparian corridors enhance movement of a forest specialist bird in fragmented tropical forest. Proc Natl Acad Sci USA 105:19774–19779
Hendrik PW (1983) Genetics of populations. Science Books International, Boston, MA
Higgins PJ, Peter JM (eds) (2002) Handbook of Australian, New Zealand and Antarctic birds. Volume 6: pardalotes to shrike-thrushes. Oxford University Press, Melbourne
Holderegger R, Wagner HH (2008) Landscape genetics. Bioscience 58:199–207
Houston P (2005) Revaluing the fringe: some findings on the value of agricultural production in Australia’s peri-urban regions. Geogr Res 43:209–223
Joshi J, Stoll P, Rusterholz HP, Schmid B, Dolt C, Baur B (2006) Small-scale experimental habitat fragmentation reduces colonization rates in species-rich grasslands. Oecologia 148:144–152
Kath J, Maron M, Dunn PK (2009) Interspecific competition and small bird diversity in an urbanizing landscape. Landsc Urban Plan 92:72–79
Koskinen MT, Hirvonen H, Landry PA, Primmer CR (2004) The benefits of increasing the number of microsatellites utilized in genetic population studies: an empirical perspective. Hereditas 141:61–67
Lada H, Thomson JR, Mac Nally R, Taylor AC (2008) Impacts of massive landscape change on a carnivorous marsupial in south-eastern Australia: inferences from landscape genetics analysis. J Appl Ecol 45:1732–1741
Landguth EL, Cushman SA (2010) CDPOP: an individual-based, cost-distance spatial population genetics model. Mol Ecol Resour 10:156–161
Legendre P, Trousellier M (1988) Aquatic heterotrophic bacteria: modeling in the presence of spatial autocorrelation. Limnol Oceanogr 33:1055–1067
Lindenmayer DB, Fisher J (2007) Tackling the habitat fragmentation pancheston. Trends Ecol Evol 22:111–166
Low Choy D (2006) Towards a regional landscape framework: is practice ahead of theory? World planning schools conference, Mexico
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
McKenzie F (1996) Beyond the suburbs: population change in the major exurban regions of Australia. Australian Government Publishing Service, Canberra
Nicholls JA, Double MC, Rowell DM, Magrath RD (2000) The evolution of cooperative and pair breeding in thorn-bills Acanthiza (Pardalotidae). J Avian Biol 31:165–176
Oksanen J, Kindt R, Legendre P, O’Hara B, Simpson GL, Solymos P, Stevens MHH, Wagner H (2009) Vegan: community ecology package for R package version 1.15-3. University of Oulu, Oulu
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pérez-Espona S, Pérez-Barberia FJ, McLeod JE, Jiggins CD, Gordon IJ, Pemberton JM (2008) Landscape features affect gene flow of Scottish Highland red deer (Cervus elephus). Mol Ecol 17:981–996
Proctor MF, McLellan BN, Strobeck C, Barclay MR (2005) Genetic analysis reveals demographic fragmentation of grizzly bears yielding vulnerably small populations. Proc R Soc B 272:2409–2416
R Development Core Team (2005) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ray N (2004) Pathmatrix, a GIS tool to compute effective distances among samples. Mol Ecol Notes 5:177–180
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rousset F (2000) Genetic differentiation between individuals. J Evol Biol 13:58–62
Segelbacher G, Cushman SA, Epperson BK, Fortin M-J, Francois O, Hardy OJ, Holderegger R, Taberlet P, Waits LP, Manel S (2010) Applications of landscape genetics in conservation biology: concepts and challenges. Conserv Genet 11:375–385
Sekercioglu CH, Ehrlich PR, Daily GC, Aygen D, Goehring D, Sandi RF (2002) Disappearance of insectivorous birds from tropical forest fragments. Proc Natl Acad Sci USA 99:263–267
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analyses. Can J Zool 69:82–90
Streiff R, Labbe T, Bacilieri R, Steinkellner H, Glössl J (1998) Within-population genetic structure in Quercus robur L. and Quercus petraea (Matt.)Liebl. assessed with isozymes and microsatellites. Mol Ecol 7:317–328
Vergara PM, Armesto JJ (2009) Responses of Chilean forest birds to anthropogenic habitat fragmentation across spatial scales. Landscape Ecol 24:25–38
Villard MA, Taylor PD (1994) Tolerance to habitat fragmentation influences the colonization of new habitat by forest birds. Oecologia 98:393–401
Vos CC, Antonisse-De Jong AG, Goedhart PW, Smulders MJM (2001) Genetic similarity as a measure for connectivity between fragmented populations of the moor frog (Rana arvalis). Heredity 86:598–608
Wilcove DS, McLellan CH, Dobson AP (1986) Habitat fragmentation in the temperate zone. In: Soulé ME (ed) Conservation Biology. Sinauer, Sunderland, pp 237–256
Wilcox BA, Murphy DD (1985) Conservation strategy—the effects of fragmentation on extinction. Am Nat 125:879–887
With KA (1994) Using fractal analysis to assess how species perceive landscape structure. Landscape Ecol 9:25–36
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
The authors would like to acknowledge CSIRO Sustainable Ecosystems (particularly Dr. Craig Miller), Birds Australia and the Wildlife Preservation Society of Australia for research funding. Birds were handled in a humane manner and measured and banded according to the requirements of Australian Bird and Bat Banding Authority. Research was authorised by the University of Queensland Animal Ethics Committee, approval number SIB/855/06/CSIRO, and Queensland Parks and Wildlife Service permit number WISP0403307. We would also like to thank two anonymous reviewers and the editor, Professor Rolf Holderegger, for insightful comments that greatly helped improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shanahan, D.F., Possingham, H.P. & Riginos, C. Models based on individual level movement predict spatial patterns of genetic relatedness for two Australian forest birds. Landscape Ecol 26, 137–148 (2011). https://doi.org/10.1007/s10980-010-9542-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-010-9542-6