Abstract
ORMOCER®s are an outstanding class of hybrid materials due to their tuneable properties, e.g. hardness, resistivity and refractive index. These materials are well-characterized with regard to their macroscopic properties, but understanding the system at the atomistic level still remains challenging. Understanding the material formation process at this level becomes especially important when three-dimensional nanoscale patterns are generated employing processes as laser-based multi-photon polymerization. We have developed an atomistic model based on the COMPASS forcefield to simulate the reference system ORMOCER®-I. We chose representative compositions for the condensation reaction product as well as for the organically cross-linked polymerized product. In the first part of the study, the results of forcefield validation experiments and the development of the atomistic model for ORMOCER®s are presented. The second part contains the results from molecular dynamics simulations at room temperature and under periodic boundary conditions, performed in order to test the feasibility of our model. The densities of the simulated materials are in very good agreement with experimentally determined densities for the unpolymerized as well as for the polymerized state, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In the last decades, organic–inorganic hybrid polymers have gained considerable interest due to their widely tunable properties and the resulting possible applications [1–3]. Such hybrid materials combine properties of pure organic polymers (e.g. flexibility and the possibility to introduce different functional groups) and of inorganic materials (e.g. hardness, stiffness, thermal and chemical stability).
An important type of class-II hybrid materials [4] are organic–inorganic hybrid polymers, e.g. ORMOCER®s (Organically Modified Ceramics; ORMOCER® is a registered trademark of Fraunhofer Gesellschaft zur Förderung der Angewandten Forschung e.V., Germany). They are synthesized in a two-step process involving firstly a condensation reaction of inorganic precursor molecules and secondly a polymerization reaction, leading to a cross-linked organic–inorganic network [1]. Various properties such as density, mechanical and chemical stability, refractive index, and electrochemical properties of the resulting material are tunable by combining different inorganic precursors and organic polymers in the hybrid systems. One of the most interesting features of many ORMOCER® systems is the possibility to pattern the hybrid polymer system, either by conventional lithography or by femtosecond laser-induced multi-photon polymerization reaction (nPP). This technique, first presented in 1997 [5], is based on a simultaneous, non-linear absorption of two or more photons in the focal volume of tightly focused laser pulses. Since the material does not interact with the laser pulses outside the focal volume, the reaction is spatially confined. In combination with suitable materials which show only small parasitic polymerization tendencies, this method allows the production of almost arbitrary three-dimensional structures in a single process by simply moving the laser focus through the material. This is one of the major improvements in comparison to conventional UV lithography, where the preparation of three-dimensional structures requires the application of layer-by-layer techniques. The non-linear absorption process in combination with a special threshold effect allows structure sizes below the diffraction limit [3, 6], which is a second advantage of multi-photon polymerization. Examples out of the large number of possible applications for the multi-photon polymerization are the generation of waveguides [7], photonic crystals [8], phase masks [9] and larger structures for medical purposes [3, 10].
ORMOCER® materials are well characterized with regard to their macroscopic properties, but understanding these systems at the atomistic level still remains challenging. Computational methods are a possible tool to overcome these restrictions. As the simulated systems need to be considerably large, force field methods are most feasible. These techniques can be used to describe systems with more than 10,000 atoms in acceptable CPU time [11]. Molecular mechanics can be used to generate different conformations and connectivities, and to compare the resulting energies. Molecular dynamics is a versatile tool to investigate the temporal evolution of the considered system. It is also possible to calculate macroscopic properties (e.g. density, Young’s modulus, radial distribution functions,…) from accurate dynamics simulations [12–14]. A first attempt to an ORMOCER® model and its analysis in force field simulations was described in 2004 [15]. An atomistic model and a molecular understanding of the processes taking place in the formation of ORMOCER® materials become especially important when very fine structures, for example with spatial resolution below 100 nm, shall be prepared and applied. In the following sections, we present the results of our approach to develop an atomistic model for the ORMOCER® archetype system ORMOCER®-I. In a preliminary step, the applicability of the chosen force field is tested in a validating study. Afterwards, the model and its application to the product after (inorganic) condensation reaction and after (organic) polymerization are presented. In this first case study, we chose the calculated and experimental densities of the different states as a test parameter to control the quality of the model employed.
2 Experimental section
All simulations were performed—as detailed further below—within Accelrys’ modeling environment ‘Materials Studio 5.0’ [16]. For the simulations, we chose the COMPASS forcefield [17], which had been validated in a preliminary modeling study of polysiloxanes [18] before and had been successfully applied to the modeling of polymer systems containing polyhedral oligomeric silsesquioxanes (POSS) [19, 20]. In addition, we performed validation studies which are closer to the system investigated here.
In order to find low energy conformations, a two-step simulation procedure was employed, consisting of a geometry optimization followed by a quenched dynamics simulation. The geometry optimization utilized the Smart algorithm implemented in the Forcite module of Materials Studio. Smart employs an optimized cascade of minimization algorithms starting with the robust steepest descent [21] method, after which an Adjusted Basis Set Newton–Raphson (ABNR) [22] minimization follows. The last step is the Quasi-Newton method in the Broyden–Fletcher–Goldfarb–Shanno (BFGS) variant [23]. The quenched dynamics simulations were performed in the canonical ensemble (NVT, constant number of particles, volume and temperature). The temperature of T = 800 K was controlled by scaling the velocities to hold the temperature in the desired region (ΔT = ±10 K). The dynamics runs lasted 10 ps with a time step of 0.1 fs. Structures were saved and energy minimized after every 100th step using the above mentioned geometry optimization procedure. For density calculations, molecular dynamics (MD) simulation in the isothermal-isobaric ensemble (NPT, constant number of particles, pressure and temperature) were performed at ambient conditions (T = 298 K, p = 0.1 MPa) applying the velocity Verlet integration algorithm [24]. The temperature during the MD runs was controlled by the Andersen thermostat [25] and the pressure by the Berendsen barostat [26]. The simulations were performed on two identical workstation systems (Intel Core i7-920, 4 × 2.66 GHz, 12 GB RAM).
3 Results and discussion
3.1 Synthetic route to ORMOCER®-I
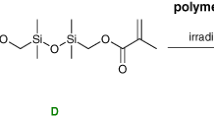
The synthesis of ORMOCER®-I follows a two-step reaction route. First, the silane precursors 3-methacryloxypropyl-trimethoxysilane (MEMO) and diphenylsilanediol (DPD), mixed in a molar ratio of 1:1, react in a condensation reaction resulting in a liquid resin (Fig. 1) [27, 28]. The condensation product consists of small building units which are the monomers for the following polymerization reaction. According to small angle X-ray scattering (SAXS) experiments, their sizes are around 1 nm (cf. Fig. S4 supporting information). In a second step, the condensation products carrying methacryloxy groups can be polymerized in a radical reaction; typically, the polymerization is initiated by light. The condensation products of the silane precursors thus act as the monomers of the polymerization reaction.
3.2 Validation of the force field
Although the applicability of the COMPASS force field for similar systems had already been shown in earlier studies [18–20], selected validation experiments were performed at the beginning of the presented modeling study. There are three characteristic segments occurring in the backbone of a typical ORMOCER® material: the Si–O–Si network, Si–O–C sites, and Si–C functionalities (Fig. 2).
For these three characteristic groups, model substances were chosen and subjected to an energy minimization procedure. The results were compared either to experimental data or to ab initio simulation results. The quality of the simulation of Si–O–C sites was checked in a study of the small silane molecules, tetramethoxysilane (TMOS) and tetraethoxysilane (TEOS). The molecular structure was constructed and a quenched dynamics simulation (NVT ensemble, T = 800 K) was performed. The resulting structures (cf. Fig. S1 supporting information for details) of the global minimum were compared to structures derived from the analysis of vibrational spectra by Ignatyev et al. [29] (TMOS) and by van der Vis et al. [30] (TEOS), respectively.
In Table 1, the bond lengths and angles found in the simulation results are compared to the experimental data. The differences of the bond lengths in the simulation range within 2 % of the experimental data, which is a good agreement for force field simulations. The differences in the resulting bond angles are somewhat larger, especially concerning the angles at the silicon atoms. The vibrational spectra revealed a slight flattening of the tetrahedron surrounding the central silicon atom for TMOS which is not visible in the simulations. This indicates that a deviation from the ideal bond angle α(OSiO) is associated with a relatively large increase in energy in the force field. This limitation of COMPASS is acceptable concerning the structural simulation of ORMOCER®s, because the small distortions of the silicon tetrahedra are assumed to have minor influence on the resulting monomer conformations and on calculated properties, e.g. the density. The experiments in both cases find S 4 symmetry [29, 30]; for TMOS, the simulation delivers the same symmetry, whereas for TEOS, simulation results in C 1 symmetry.
To prove whether the force field describes the Si–O–Si network correctly, five silicic acid oligomers were simulated applying the COMPASS force field. These oligomers are a four-membered ring, a double-four-membered ring, a five-membered ring, a six-membered ring and a double-six-membered ring (cf. Fig. S2 supporting information for structures).
The comparison of the resulting bond lengths of the four-membered and the double-four-membered ring to ab initio results from Xue et al. [31] and Pereira et al. [32] (Table 2) again show a very high degree of correspondence. The bond angles are in good agreement except for the [SiO4] tetrahedra, where the ab initio results show a slight distortion which is not reproduced in the force field simulations.
The bond length and angles of a silicic acid six-membered ring resulting from force field simulations are given in Table 3 in comparison to the ab initio results from Pereira et al. [33]. The data of both calculations are in good agreement. A slight distortion of the [SiO4] tetrahedra is observed in the ab initio simulation of a six-membered ring only. Additionally, this molecule shows tilted silicon tetrahedra resulting in optimized intramolecular interactions, especially the formation of hydrogen bonds. This is a restriction of the simulation of single molecules in vacuo, it is also reported in the ab initio study of Pereira et al. Therefore, the structures and energies of the simulated ORMOCER® monomers may also be influenced by the named limitations of simulating individual molecules in vacuo. We therefore decided to not investigate further single molecules for validation, but to focus on larger systems, where intermolecular interactions come into play.
In particular, the investigation of several precursor molecules or of sets of monomers in simulated unit cells under the application of periodic boundary conditions (PBCs) appeared promising. The crystal structures of diphenylsilanediol [34] (DPD), one of the precursor molecules for the preparation of ORMOCER®-I, and of octaphenylcyclotetrasiloxane [35], the cyclic tetramer of DPD obtained by condensation reaction, are known from single crystal analysis. Therefore, unit cells containing these molecules were simulated using the COMPASS force field with applied PBCs in a quenched dynamics simulation.
The overlay of the structure of the diphenylsilanediol molecules resulting from our simulation and the reported [34] crystal structure in Fig. 3 (right picture) reveals the very good congruency. The length of the unit cell is a little shortened in all three directions in the simulation (cf. Table S1 supporting information for complete cell parameters). This was expected, because the geometry optimization step in the quenched dynamics part is performed at T = 0 K. This negligence of thermal effects leads to slightly decreased distances between the DPD molecules resulting in a slightly reduced size of the unit cell. Apart from this anticipated effect, the molecular and the crystal structure are well reproduced in the force field simulation. Even the inversion center determined in the experimental study is present also in the simulation result, although that symmetry element was not included in the input structure (the simulation was carried out in space group P1).
Crystal structure of diphenylsilanediol. Left as published by Fawcett et al. [34]; middle energy-optimized structure (PBCs applied); right superposition of experimental (red) and simulated (blue) structure. The superposition shows a good agreement of the simulation result with the experimentally determined structure (Color figure online)
Figure 4 shows the crystal structure of octaphenylcyclo-tetrasiloxane. The superposition of the simulated structure and the reported one [35] reveals a high degree of correspondence between the structures of the cyclic molecules. Due to the negligence of thermal effects, the distance between the two vertical rows (marked with dashed lines in Fig. 4) of molecules is shorter in the simulated structure, leading to a slight shift of the rows against each other. This results in smaller unit cell lengths and an enlarged β angle (cf. Table S2 supporting information for complete cell parameters). The space group obtained in the simulation is P21/c, as reported in the single crystal structure analysis [35], although the simulation was performed in space group P1.
Crystal structure of octaphenylcyclotetrasiloxane. Left as published by Hossain et al. [35]; middle energy-minimized structure (PBCs applied); right super-position of experimental (red) and simulated (blue) structure. The superposition shows a good agreement of the simulation result with the experimentally determined structure (Color figure online)
All results from the validation experiments show a high correspondence of the simulated structures with already published ones, which were derived either in ab initio simulations or experimentally. Bond lengths are very well reproduced, bond angles show only small deviations and the applicability of PBCs is also confirmed. The COMPASS force field should therefore be well-suited for the simulation of ORMOCER®s.
3.3 Condensation products—monomers
For the modeling of ORMOCER®-I, a procedure similar to the synthesis route was developed. In analogy to the products of the condensation reaction, oligosiloxanes were created. Small angle X-ray scattering experiments had revealed structure sizes of around 1 nm for the Si–O backbone of these condensation products (cf. Fig. S4). We therefore assumed all monomer structures as valid which fulfil this size condition (these typically contain 4–6 Si atoms) and which contain the silane building blocks 3-methacryloxypropylsilyl (M, derived from MEMO) and diphenylsilyloxy (D, derived from DPD) in a ratio of 1:1 (or in a ratio near to that). Valid monomer molecules were constructed and collected in a database. For example, one group of possible monomers are linear chains consisting of a total number of six silane building units, three D and three M units. The scheme in Fig. 5 shows all ten possible precursor sequences for six-membered chains that satisfy the two given requirements of size and composition. In addition to the six-membered linear chains described above, linear chains with four and five silane building units, branched chains containing five or six building units as well as cyclic monomers containing four-membered rings were also evaluated as possible structures. The cyclic product of a self-condensation reaction of four DPD molecules, i.e. octaphenylcyclotetrasiloxane, was considered as a monomer, too, as it was found to be present in the liquid resin in silicon nuclear magnetic resonance experiments (29Si-NMR, signal at around −43 ppm) [27, 36].
The different monomer molecules were constructed by connecting the energy-minimized precursor units. These molecules were then investigated by a geometry optimization approach utilizing the Smart algorithm implemented in the Forcite Module of Materials Studio [16]. Depending on the initial geometry, different low-energy structures were found for the same monomer model which exhibited only small differences in energy. These results gave a first insight into the system: the monomer units seem to be quite flexible and can adopt different nearly equivalent geometries corresponding to different local minima. A simple geometry optimization procedure is obviously not suitable to find the global minimum. Therefore, a quenched dynamics approach was used with the intent to overcome the drawbacks of the geometry optimization procedure. For each monomer, the resulting structure from the geometry optimization was used as the starting point. The quenched dynamics (NVT, T = 800 K) run revealed that structures with considerably lower energies can be found. However, again the monomers can adopt different geometries with quite similar energies, as the quenched dynamics simulation result in Fig. 6 shows exemplarily. The minimum structures of all monomers were collected in a database and used for further investigation. Since the monomers are highly flexible—even at room temperature a lot of energetically similar conformations can be adopted—the calculated energies were of minor interest. The lowest energy conformation was therefore only used as a defined starting point for further investigations.
Example result of a quenched dynamics simulation (NVT ensemble, T = 800 K) of an ORMOCER®-I monomer model (linear six-membered chain No. 3 in Fig. 5). The minimum structure was found at frame number 665 with a total potential energy of −297.00 kcal mol−1. Other conformations have similarly low energies
In order to be able to evaluate intermolecular interactions between the monomers, simulation cells containing four deliberately selected, energy-minimized monomers were constructed, corresponding to step A in Fig. 7. The amount of four monomers per unit cell was chosen as a good compromise between sufficiently large simulation system size and acceptable requirement of CPU time.
Overview of the modeling process for ORMOCER®-I. Clockwise, starting in upper left Four isolated monomers are used to construct a simulation cell (step A) which then undergoes a quenched dynamics (QD) run (step B, NVT, T = 800 K); In this model, possible sites for polymerization reactions are identified according to distance criterion (step C) and bonds are inserted there (step D). Finally, a quenched dynamics run is carried out on the polymerized material (step E, NVT, T = 800 K)
In total, 28 simulation cells with different compositions of monomers (cf. Tables S3 supporting information for cell compositions) were constructed in this way, to cover a broad range of compositions and system sizes in order to sample the characteristics of an amorphous hybrid system as ORMOCER®-I. To receive suitable structures for further investigation, NVT (800 K) quenched dynamics simulations applying periodic boundary conditions (PBCs) were performed (step B in Fig. 7). The structure with the lowest energy served as the starting point for an NPT molecular dynamics (MD) simulation at ambient conditions. The MD simulation time was set to 100 ps. The density was calculated at every 100th step of the MD simulation. Figure 8 shows a typical evolution of the density with time during the dynamics simulation of such a simulation cell (entry 2 in Table 4, see below).
Density evolution during a 100 ps NPT MD simulation of an example ORMOCER®-I simulation cell (entry 2 in Table 4) containing four monomers at ambient conditions
In the beginning of the simulation time, the density decreases until an equilibrium state is reached (approximately after 40 ps) where the density oscillates around an average value. Since the simulation of the other models results in a similar behavior, 50 ps appear well suited to equilibrate the system. The densities of all 28 simulation cells were thus calculated by averaging the calculated densities over the second 50 ps of the simulation time of the room temperature MD simulation (every 100th step). In selected model systems the simulation time was extended to 150 ps, but the inclusion of the additional 50 ps in the calculation of the densities did not lead to significant deviations (cf. Table S5 supporting information for results).
As listed in Table 4, the densities ρbp (bp: before polymerization), calculated as mentioned above, cover a range of densities from 1.14 to 1.21 g cm−3, located around the experimental value of 1.175 g cm−3 for the monomer resin. Nevertheless, some of the monomer compositions are obviously better suited to describe the characteristics of the liquid resin of ORMOCER®-I than others. In general, those simulation cells which contain the cyclic tetramer of DPD, the octaphenylcyclotetrasiloxane cyclo-DDDD (a species which cannot undergo polymerization), exhibit densities that are closer to the experimental value (entries 21–24 and 26–28 in Table 4). Noteworthy, the cyclo-DDDD monomer was found to be a prominent species in the product of the condensation reaction by 29Si-NMR experiments (signal at around −43 ppm) [27, 36].
3.4 Polymerization
The polymerization reaction involves the formation of new bonds, a process which is out of scope of this type of force field simulations. To circumvent this limitation, the monomers in the optimized simulation cells were connected manually instead according to the following procedure: First, the distances between candidate 3-methacryloxypropyl groups were measured (step C in Fig. 7), second, the bonds were then inserted between pairs of candidate groups that are sufficiently close to each other (step D in Fig. 7). We took into account the results of μ-RAMAN spectroscopy experiments on polymerized ORMOCER®-I which revealed a degree of polymerization (PD) of approximately 60 % [37]. Hence, the simulated polymerization process was guided to obtain a similar PD (50–72 %). The simulation cells, now containing the polymerized units, were again energy-optimized (step E in Fig. 7), and the change of the density with time was analyzed in the same way as mentioned before. Again, an equilibration time of 50 ps was shown to be suitable; densities of the materials after polymerization, ρap, were then calculated as the average of the density values of the second 50 ps of simulation time. These densities are also listed in Table 4. They can furthermore be compared to the experimental value for polymerized ORMOCER®-I. After polymerization and simulation, the densities of the simulation cells (1.17–1.22 g cm−3) again are close to the experimental value of 1.22 g cm−3 of polymerized ORMOCER®-I (deviations are smaller than 5 % in most of the cases), as it was the case for the non-polymerized condensation product, although in most of the cases the density is slightly underestimated. Again, the simulation cells containing the octaphenylcyclotetrasiloxane cyclo-DDDD are found to have densities closer to the experimental value. The largest deviations (cells 26 and 27) are most likely caused by the low PD in these model structures. It had to be adjusted to 50 %, because the next possible value would have been 75 % due to the small total number of polymerizable groups in these simulation cells.
It is noteworthy that—notwithstanding the deviations of the calculated densities ρbp from the experimental values—the density increase upon polymerization Δρ (which corresponds to the polymerization shrinkage) is in many cases well reflected in the results. The experimental value is 3.8 %, the calculated ones range from −1.9 to 4.1 %.
4 Conclusions
Here, we present the first in-depth computational study of an organic–inorganic hybrid material with a very complicated composition and a complex formation process. It is shown, that the COMPASS force field is applicable for the modeling of ORMOCER® materials. In contrast to other systems, the complexity is not only attributed to the large number of atoms which have to be taken into account and to the high flexibility of the polymers (as in the case of simulation of biopolymers, e.g. proteins). It is in addition increased by the ambiguity of the molecular composition. The presented atomistic model delivers first hints for possible structure candidates and compositions in the reference system ORMOCER®-I. This model follows the preparation route for ORMOCER® materials. Its cornerstones are the silane precursors, their condensation products (monomers), ensembles of monomers (representing the liquid resin) and polymerized condensation products (representing the final hybrid material). The simulation strategy consists of geometry optimization and quenched dynamics simulation steps. Since a polymerization reaction cannot be modeled with force field methods, an alternative procedure was applied to introduce the new connections manually.
Both techniques, i.e. the optimization procedure and the manual polymerization were successfully tested in modeling studies of monomer ensembles. Each ensemble contained four ORMOCER®-I monomers chosen deliberately from the monomer database. The generated models were investigated in terms of molecular dynamics simulations at room temperature for the non-polymerized as well as for the polymerized compound. As judged by the high degree of correspondence between the experimental densities and the simulation results, the assumed model is apparently well-suited to favourably represent ORMOCER®-I. The agreement was improved in those cases where the monomer cyclo-DDDD (octaphenylcyclotetrasiloxane) was present in the simulated ensembles. This is noteworthy as this species was shown to be a prominent species in the liquid resin (29Si-NMR) [27, 36].
In the future, the dependence of the results on the size of the simulated systems will be investigated. For this purpose, the generation and simulation of supercells of the existing simulation cells and of larger simulation cells is planned. Furthermore, we will try to enhance our work by making possible comparisons with the results from other experimental methods. Also, in order to test the transferability of our model, other ORMOCER® systems will be simulated.
References
Haas K-H, Rose K (2003) Rev Adv Mater Sci 5:47
Haas K-H, Wolter H (1999) Curr Opin Solid State Mater Sci 4:571
Houbertz R, Steenhusen S, Stiche T, Sextl G (2010) In: Duarte FJ (ed) Coherence and ultrashort pulse laser emission. InTech, Rijeka
Kickelbick G (2006) Hybrid materials, synthesis, characterization, and applications. Wiley-VCH, Weinheim
Maruo S, Nakamura O, Kawata S (1997) Opt Lett 22:132
Tanaka T, Sun HB, Kawata S (2002) Appl Phys Lett 80:312
Schmidt V, Kuna L, Satzinger V, Houbertz R, Jakopic G, Leising G (2007) Proc SPIE 6476:P4700
Houbertz R, Declerck P, Passinger S, Ovsianikov A, Serbin J, Chichkov BN (2008) In: Wehrspohn RB, Kitzerow H-S, Busch K (eds) Nanophotonic materials. Wiley-VCH, Weinheim
Jia BH, Serbin J, Kim H, Lee B, Li JF, Gu M (2007) Appl Phys Lett 90:073053
Claeyssens F, Hasan EA, Gaidukeviviute A, Achilleos DS, Ranella A, Reinhardt C, Ovsianikov A, Xiao S, Fotakis C, Vamavakaki M, Chichkov BN, Farsari M (2009) Langmuir 25:3219
Kunz RW (1997) Molecular modelling für anwender. Teubner, Stuttgart
Patel RR, Mohanraj R, Pittman CU Jr (1996) J Polym Sci, Part B: Polym Phys 44:234
Economou IG, Garrido NM, Makrodimitri ZA (2010) Fluid Phase Equilib 296:125
Canales M, Curcó D, Alemán C (2010) J Phys Chem B 114:9771
Kahlenberg F (2004) Ph.D. Thesis, Julius-Maximilians-Universität Würzburg
Materials studio, version 5.0. Accelrys Software Inc., San Diego (2009)
Sun H (1998) J Phys Chem B 102:7338
Sun H, Rigby D (1997) Spectrochim Acta, Part A 53:1301
Bharadwaj RK, Berry RJ, Farmer BL (2000) Polymer 41:7209
Bizet S, Galy J, Gérard J-F (2006) Polymer 47:8219
Wiberg KB (1965) J Am Chem Soc 87:1070
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) J Comput Chem 4:187
Shanno DF (1970) Math Comput 24:647
Swope WC, Andersen HC, Berens PH, Wilson KR (1982) J Chem Phys 76:637
Andersen HC (1980) J Chem Phys 72:2384
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) J Chem Phys 81:3684
Buestrich R, Kahlenberg F, Popall M, Dannberg P, Müller-Fiedler R, Rösch O (2001) J Sol Gel Sci Technol 20:181
Houbertz R, Domann G, Cronauer C, Schmitt A, Martin H, Park J-U, Fröhlich L, Buestrich R, Popall M, Streppel U, Dannberg P, Wächter C, Bräuer A (2003) Thin Solid Films 442:194
Ignatyev IS, Lazarev AN, Tenisheva TF, Shegolev BF (1991) J Mol Struct 244:193
van der Vis MGM, Konings RJM, Oskam A, Snoeck TL (1992) J Mol Struct 274:47
Xue X, Kanzaki M (1998) Phys Chem Miner 26:14
Pereira JCG, Catlow CRA, Price GD (1999) J Phys Chem A 103:3252
Pereira JCG, Catlow CRA, Price GD (1999) J Phys Chem A 103:3268
Fawcett JK, Camerman N, Camerman A (1977) Can J Chem 55:3631
Hossain MA, Hursthouse MB, Malik KMA (1979) Acta Crystallogr Sect B: Struct Sci 35:522
Hoebbel D, Reinert T, Schmidt H (1996) J Sol Gel Sci Technol 7:217
von Landgraf F (2010) Dipl. Thesis, Fraunhofer ISC, Würzburg
Acknowledgments
The financial support of the Deutsche Forschungsgemeinschaft (DFG) within the priority program 1327 “sub-100 nm structures” is gratefully acknowledged. The authors would like to thank the cooperation partners F. Burmeister, S. Nolte, A. Tünnermann and U.D. Zeitner at the Friedrich-Schiller University of Jena for fruitful discussion.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fessel, S., Schneider, A.M., Steenhusen, S. et al. Towards an atomistic model for ORMOCER®-I: application of forcefield methods. J Sol-Gel Sci Technol 63, 356–365 (2012). https://doi.org/10.1007/s10971-012-2794-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10971-012-2794-7