Abstract
In this paper, three mutants from wild Saccharomyces cerevisiae HBU2.558, called U2.558, UN2.558, and UNA2.558, were screened by UV, sodium nitrite, Atmospheric and room temperature plasma, respectively. Glutathione production of the three mutants increased by 41.86, 72.09 and 56.76%, respectively. We detected the activity of glutathione synthetases and found that its activity was improved. Amino acid sequences of three mutant colonies were compared with HBU2.558. Four mutants: Leu51→Pro51 (L51P), Glu62→Val62 (E62V), Ala332→Glu332 (A332E) and Ser653→Gly653 (S653G) were found in the analysis of γ-glutamylcysteine ligase. L51 is located adjacently to the two active sites of GCL/E/Mg2+/ADP complex in the overall GCL structure. L51P mutant spread distortion on the β-sheet due to the fact that the φ was changed from −50.4° to −40.2°. A mutant Leu54→Pro54 (L54P) was found in the analysis of glutathione synthetase, and L54 was an amino acid located between an α-helix and a β-sheet. The results confirm that introduction of proline located at the middle of the β-sheet or at the N- or C-terminal between α-helix and β-sheet or, i.e., L51P and L54P, changed the φ, rigidity, hydrophobicity and conformational entropy, thus increased protein stability and improved the enzyme activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Glutathione (γ-glutamylcysteinylglycine; GSH) is an intracellular low molecular-weight tri-peptide which is made up of glutamic acid, cysteine and glycine [1]. It occurs in all biological cells, whose content is higher in yeast and animal, is less in the plant. GSH has multiple physiological activities, such as anti-oxidization, amino acid transport, detoxification and immune, etc. Because of the powerful activity, GSH is widely used as an ingredient in cosmetics, food and pharmaceutical.
Traditional methods of microbial mutation breeding included physical mutation methods (e.g., γ-rays, X-rays, UV light, particle radiation, etc.) and chemical mutagenesis methods (e.g., alkylating agents, azides, diethyl sulphate and ethyl methanesulphonate, etc.) [2, 3]. Recently, atmospheric and room temperature plasma (ARTP), a robust, novel, and environmental friendship mutagenesis method have been applied to physiological mutagenesis of microorganisms in microbial breeding [4]. Wang et al. [5] employed the ARTP mutagenesis method to Arthrobacter so as to produce dextranase, and then two mutants were obtained. The dextranase activity increased by 19 and 30%. Li et al. [4] obtained mutants with higher arachidonic acid (ARA) production from Mortierella alpine combined with ARTP and diethylsulfate treatments. ARTP mutation had been well applied to generate multiple mutants in fungi [4], bacteria [6] and Spirulina [7]. But in the S. cerevisiae, this method has not been applied to the high generation of GSH mutants yet.
In yeast, two ATP-dependent sequential reactions are took out to synthesize GSH: the first and primary rate-limiting reaction is catalyzed by γ-glutamylcysteine ligase (GCL/Gsh1; encoded by GSH1) to ligate l-glutamate and l-cysteine, to generate a γ-glutamylcysteine (γ-GC), and the second reaction is catalyzed by glutathione synthetase (GSS/Gsh2; encode by GSH2) in the pathway [8,9,10]. According to the Jozefczak [11], GCL is rate-limiting enzyme because of the three reasons: firstly, GSH is feedback inhibition of GCL; secondly, de novo synthesis GCL as well as GSS; finally, GCL activity is regulated via post-translational redox controls.
2 Materials and Methods
2.1 Yeast Strains and Medium
The initial S. cerevisiae strain HBU2.558 was provided by Institute of Microbiology of Chinese Academy of Sciences (China) and preserved in our laboratory, which can produce 2.5 mg glutathione by gram of the waterish cells. The seed medium was yeast extract-peptone-dextrose (YPD) medium containing 1% yeast extract, 2% peptone, and 2% glucose, pH7.0. Screening culture medium was YPD agar medium. GSH fermentation medium contains 0.7% (NH4)2SO4, 0.7% KH2PO4, 0.6% K2SO4, and 0.2% MgSO4.
2.2 Mutagenesis Protocol
The tested strain was grown in YPD medium overnight at 30 °C with mechanical shaking (200 rpm). Adjusting the concentration of yeast to OD600 = 0.6 with sterile saline, cells were harvested by centrifugation. The S. cerevisiae strain treatment with the UV irradiations, sodium nitrite and plasma jet from ARTP was based on the methods described by Ramzan et al. [12], Routledge et al. [13] and Wang et al. [5], respectively. Lethality rate (%) was calculated after counting the individual control colonies and survival mutants.
2.3 GSH Analysis
The tested strains were cultured in YPD medium (50 mL) overnight at 30 °C with mechanical shaking (200 rpm). After the step, the cells (~2 × 107cells/mL) were transferred to fermentation medium (150 mL) for 2 days, and cells were collected by centrifugation. GSH was extracted according to the method of Nakamura et al. [14]. GSH concentration in the supernatant was measured immediately using high-performance liquid chromatography (HPLC) system (Class L-2000 system, Hitachi, Japan) with C18 reverse chromatographic column (250 × 4.6 mm, particle size 5 µm, Thermo, North America) according to the method of Gales [15]. Derivative materials were detected with an ultraviolet detector (wavelength 412 nm).
2.4 In vivo Enzymatic Assay
Cells were broken by Ultrasonic cell crushing system (650 W, JY92-II, Rongyan, China) and cytosol was extracted with KH2PO4–K2HPO4 buffer (50 mM, pH 8.0). One unit of enzymatic activity is defined as the amount that catalyzes 1 μmol of product per min. The in vitro enzymatic activity of glutathione synthetases was determined according to the method described by Toroser and Sohal [16] with a slight modification. Briefly, 4.0 mL pre-warmed of GSH reaction medium containing 60 mM l-glutamate, 20 mM l-Cys, 40 mM l-Gly, 5 mM ATP, 20 mM MgCl2 and 50 mM KH2PO4–NaOH buffer (50 mM, pH 7.5) was pre-incubated for 5 min at 40 °C. The enzyme reaction was initiated at 40 °C by addition of 1.0 mL of cytosol for 60 min. An equal volume of KH2PO4–K2HPO4 buffer instead of cytosol was added to the reference cuvette as a blank control. The supernatant was harvested by centrifugation (at 4 °C, 12,000 rpm for 10 min) and refiltered through 0.25 μm PTFE Acrodisc syringe filters. GSH production in the supernatant was measured using HPLC system immediately or stored at −70 °C for no longer than 24 h before analysis.
2.5 Detection of Mutations
DNA isolation, analysis and cloning were based on the Molecular cloning: a laboratory manual [17]. Two pairs of primers, Gsh1 and Gsh2, were designed based on the sequences of Gene GSH1 and GSH2 from S. cerevisiae strain of S288c (http://www.yeastgenome.org). GSH1 was amplified using the primers Gsh1f 5′-TCTTATGAGTGGAGCAATCG-3′ and primer Gsh1r 5′-TTCAATCACCGTGTCACC-3′. GSH2 was amplified using the primers Gsh2f 5′-CCAAAGGTAGCAAAGTGCC-3′ and Gsh2r 5′-TATGTTTCGGTATGTCTGCC-3′. The PCR program was initiated at 94 °C for 5 min, followed by 30 cycles of 94 °C for 45 s, 54 °C for 45 s, 72 °C for 1.5 min, and a final extension an 72 °C for 10 min. The amplification products were analyzed by 1.5% agarose gel electrophoresis, and purified with the DNA Gel Extraction Kit (Takara, Japan) and then cloned into pMD18-T (Takara, Japan). Sequence reactions were carried out by Aokesw (China). Amino acid sequences of three different GCL and GSS were compared with wild-type sequences in a multiple sequence alignment.
3 Results
3.1 Lethality Rate of S. cerevisiae from Mutation
Mutagenesis results in a dose-dependent lethality of Saccharomyces cerevisiae relative to the treatment time. The lethality rose to 50, 80, 87 and 91.2% respectively, after cells were treated with UV irradiation for 60, 120, 180 and 240 s. The lethality rose to 40.1, 65.7, 75.6 and 81.2% respectively, after cells were treated with the sodium nitrite for 90, 150, 210 and 270 s. The lethality rose to 78.8, 92.3, 96.3 and 98.5% respectively, after cells were treated with the plasma jet for 60, 120, 180 and 240 s. According to earlier literature on mutagenesis breeding, the mortality rate of 80% was considered appropriate. Therefore, the time for the treatment of S. cerevisiae was primarily determined to be 2.0, 4.5 and 1.5 min by UV, sodium nitrite and ARTP, respectively.
3.2 Mutagenesis
UV mutation, Nitrite-induced mutations and ARTP were carried out in this study. Starting with strain HBU2.558, 200 colonies were selected by UV mutant. After screening and three generations of culture, strains U2.558 still maintained high GSH-producing ability. Starting with mutant strain U2.558, a second treatment of Nitrite-induced mutant was carried out, and 200 colonies were selected. Among these 200 colonies, 57 positive mutant strains were obtained. One positive mutant strain, here named UN2.558 was selected. Like this, another treatment of ARTP was carried out, the survival strains were cultivated, then, a high positive (about 61%) mutation rates were accomplished. Finally, strain UNA2.558 was obtained, whose GSH-production ability was the highest of this round mutants.
3.3 GSH Content and In Vivo Enzymatic Activity
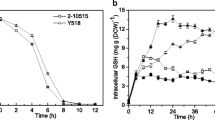
The strains used in this study were listed in Table 1, and the content of GSH determined by HPLC system was shown at Fig. 1. The retention time of GSH standard was 8.37 min, and the curve of standard is: A = 85887C + 1897. A is the areas of GSH determination by HPLC system, C is the content of GSH produced by strain. The results showed that GSH production of the three mutants increased by 41.86, 72.09 and 56.76% compared with parent strain.
The GSH content of three mutants increased sustainable showed that the enzymatic activity of glutathione synthetases were likely to changed. In comparison, the mutant U2.558, UN2.558 and UNA2.558 showed a higher activity than their parent strain HBU2.558 (~112, ~151 and ~167% of parent strain activity respectively).
3.4 Comparison of the GCL and GSS Sequence Between Mutants and Wild-type Strain
In order to found out the reasons why the activity of glutathione synthetases was improved, the GSH1 and GSH2 genes were cloned. Full-length DNA clones of 2270 and 1780 bp were obtained respectively as expected (Fig. 2). The sequence of GCL from HBU2.558 strain can encode a polypeptide of 678 amino acids and the protein molecular weight was 78.253KD, the sequence of GSS can encode a polypeptide of 491 amino acids and the protein molecular weight was 55.815 kDa.
3.4.1 Variation of GCL
The results of the multiple sequence alignment shows that three different mutants shows 99.92, 99.92 and 99.85% similarity to that of the HBU2.558, respectively. Two forms of significant base substitutions, C→T (account for 40% of total mutations) and C(G)→A were led by UV irradiations. Two base substitutions : C→T and T→C were leaded by sodium nitrite, and plasma jet from ARTP were induced only one base mutant: T→C.
The GCL crystal model of HBU2.558 was built by Swiss-model (http://swissmodel.expasy.org/interactive) and Viewerlite software (Fig. 3a) based on S. cerevisiae S288c crystal structures. The GCL structural fold of HBU2.558 shows 99.85% similarity to that of the S288c. Major differences between the analyzed GCL were found in positions 51and 62, which are positioned near the glutamate binding pocket, especially Leu51 is located adjacent to the active site of the GCL/E/Mg2+complex (Fig. 3b). UV irradiations were induced into three mutants: Glu62→Val62 (E62V), Ala332→Glu332 (A332E) and Ser653→Gly653 (S653G) of amino acids sequence of GCL. No mutant was found by sodium nitrite, and Leu51→Pro51 (L51P) was induced by plasma jet.
a Ribbon representation of the recombinant GCL crystal structure of HBU2.558 built by Swiss-model online software and ViewerLite software. A functionally active GCL monomer (78.28KD) is contained in the asymmetric unit (center). The catalytic domain comprise of six anti-parallel β-strands surrounded by α-helices. The N terminus of the sequence considered is met and it defines a lip of the active site, which is situated in a central cavity of the protein. The core β-sheet is colored in blue, α-helical elements in red, and loop regions in gray. The differences between the analyzed GCL were shown in ball and stick. Leu 51 was colored in yellow, which was positioned on a β-sheet, Glu 62 was colored in green, Ala 332 was in purple, and Ser 653 was in red. b Multiple sequence alignment of GCL. The picture was generated by ClustalW Multiple alighment of sequence using the BioEdit software. Four mutants: L51P, E62V, A332E and S653G were found from GCL analysis. (Color figure online)
The kinds of Physical parameter such as the theoretical pI, instability index (II), and Grand average hydropathicity (GRAVY) were analyzed by Protparam online software (http://web.expasy.org/protparam/). Compared with HBU2.558, the heoretical pI of U2.558 and UN2.558 were unchanged, but the physical parameter such as II and GRAVY of these two strains were changed from 42.71 to 42.06 and from −0.511 to −0.507 (Table 1). Thus, the structure of GCL from U2.558 and UN2.558 was more stable than that from HBU2.558, and the thydrophilicity tended to increased. But for the UNA2.558, II and GRAVY were 42.6 and −0.515, the two dates were between the HBU2.558 and U2.558.
3.4.2 Variation of GSS
UV irradiations led to a base mutant, C→A. Three base mutants: T→C (50%), C→T (25%) and G→T (25%) were induced by sodium nitrite and two base substitutions forms: C→T and T→G were by plasma jet from ARTP. As to amino acid sequence, the differences between the analyzed GSS were found, Leu54→Pro54, at the position 54 induce by sodium nitrite, which is positioned between a α-helix and β-sheet (Fig. 4).
4 Discussion
Microbial breeding by altering the genomes is of great importance for biotechnology research and bio-industry [3]. Since Stadler et al. [18] reported that X-ray could be used in the mutagenesis of maize, various artificial mutagenesis methods for improving mutation rates have been developed and applied in industrial production. Like a person’s signature, a mutagen signature is unique and cognizable to its owner, because it is distinguished from others. For UV breeding, two pyrimidines (C or T) were adjacent and revealed the unique UV-induced CC→TT or C→T substitutions at dipyrimidine sites substitutions [19]. Routledge et al. [13] reported that the most commonly induces by nitrite were GC→AT transitions, followed by GC→TA transversions. The main factor affecting DNA is the chemical-reactive species, such as helium lines, oxygen atom lines, N2 lines, and hydroxyl (OH) molecular band, which destroyed mononucleotides’ phosphates group and then broke oligonucleotides into fragments. Therefore, helium RF APGD plasma jet is more energetic and mass than UV irradiations. The results of the multiple sequence alignment were consist with the above reports approximately. But transversions were not found because of the tested strains were different (S. cerevisiae cells VS human AD293 cells) and pHs of sodium nitrite were used possibly. Based on the Table 1, ARTP mutant method can be regarded as a potential substitute for S. cerevisiae screening. And alternating physical and chemical mutation methods gave the better mutagenesis more than a single method of mutation.
GSH deficiency affects various systems of the human body and the growth rate of microorganism. Ohtake et al. [20] reported that the growth rate of the mutants, which were GSH biosynthesis-deficient, were much slower than the parent strains. And the growth rate of the mutants were restored after GSH was added to the medium. In this study, the colonies selected grow faster and bigger relatively than other colonies in the screening medium. The result shows that the GSH produced by smaller colonies was really lower than bigger’s, but not every bigger colony can produce higher content GSH.
In the present experiment, Toroser and Sohal’s [16] method is used to test the enzymatic activity of GCL and GSS, which can’t directly detect the activity of GCL and GSS separately, but their mutation site of the three mutants are coincidently complementary so as to suggest the activity change of GCL and GSS: Compared with the original strains, the enzymatic activity of mutant U2.558 increased by 11.7%, amino acid sequence of GSS didn’t change, substitution (E62V, A332E, S653G) happened to the amino acid in three different positions, which suggests that the GCL enzymatic activity of mutant U2.558 increased. Compared with the mutant U2.558, the enzymatic activity of mutant UN2.558 increased by 51%, amino acid sequence of GCL didn’t change, and one amino acid Leu54 of GSS was replaced by Pro54, which means that this site mutation of GSS could dramatically increase enzymatic activity. Compared with the mutant UN2.558, the enzymatic activity of mutant UNA2.558 increased by 74.5%, amino acid sequence of GSS didn’t change, and one amino acid Leu51 of GCL was replaced by Pro51, which means that this site mutation of GCL could dramatically increase enzymatic activity.
The structure of GCL comprises residues 2-678, ADP and three magnesium ions (Mg2+). It has a core β-sheet comprising with a catalytic domain (residues 18-387 and 442-518) and a small domain (residues 1-16 and 388-441). Three bound metal ions, Mg2+were identified in the GCL/E50/ Mg2+/ADP complex, GCL/E52/Mg2+/ADP complex and GCL/E103/ Mg2+/ADP complex respectively [21]. In the GCL/E52/Mg2+/ADP complex, ADP is largely solvent-exposed, with its β-phosphate positioned near the γ-carboxylate of the bound glutamate substrate and forming a β-sheet.L51 is the neighbor of the two GCL/E/Mg2+/ADP complex in the overall GCL structure. Proline is more rigid than other naturally occurring amino acids and, in proteins, lacks an amide hydrogen [22]. L51P mutant spread distortion on the β-sheet due to the fact that φ was changed from −50.4° to −40.2°. This inferred that introduction of proline located at the middle of the β-sheet may has a steric effect and then affecting the structure–function relationship of protein.
The structure of GSS is composed of a large “main” domain and a small “lid” domain. The main domain is characterized by an α/β-fold comprised of two topological sub-domains [23]. As a helix-breaker, proline is fixed at the first turn of an α-helix expected to increases the internal hydrophobicity and decrease the conformational entropy of the denatured domain, therefore to lead to the α-helix stabilization and consequently stabilizing the protein [22, 24]. Overall, the paper results confirm that introduction of proline located at the middle of the β-sheet or at the N- or C-terminal between α-helix and β-sheet or, i.e., L51P and L54P, changed the φ, rigidity, hydrophobicity and conformational entropy, thus increased protein stability and improved the enzyme activity.
Abbreviations
- S .cerevisiae :
-
Saccharomyces cerevisiae
- GSH:
-
Glutathione
- ARTP:
-
Room temperature plasma
- GCL:
-
γ-Glutamylcysteine ligase
- GSS:
-
Glutathione synthetase
- YPD:
-
Yeast extract-peptone-dextrose
References
Circu ML, Aw TY (2012) Glutathione and modulation of cell apoptosis. Biochim Biophys Acta 1823:1767–1777
Lu Y, Wang LY, Ma K., Li G, Zhang C, Zhao HX, Lai QH, Li HP, Xing XH (2011) Characteristics of hydrogen production of an Enterobacter aerogenes mutant generated by a new atmospheric and room temperature plasma (ARTP). Biochem Eng J 55:17–22
Zhang X, Zhang XF, Li HP, Zhang C, Zhou QQ, Wang LY, Chang HB, Oda Y, Xing XH (2014) Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl Microbiol Biotechnol 98:5387–5396
Li XY, Liu RJ, Li J, Chang M, Liu YF, Jin QZ, Wang XG (2015) Enhanced arachidonic acid production from Mortierella alpine combining atmospheric and room temperature plasma (ARTP) and diethyl sulfate treatments. Bioresour Technol 177:134–140
Wang XB, Lu MS, Wang SJ, Fang YW, Wang DL, Ren W, Zhao GM (2014) The atmospheric and room-temperature plasma (ARTP) method on the dextranase activity and structure. Int J Biol Macromol 70:284–291
Wang Q, Feng LR, Wei L, Li HG, Wang L, Zhou Y, YU XB (2014) Mutation breeding of lycopene-producing strain Blakeslea Trisporaby a novel atmospheric and room temperature plasma (ARTP). Appl Biochem Biotechnol 174:452–460
Fang MY, Jin LH, Zhang C, Tan YY, Jiang PX, Ge N, Li HP, Xing XH (2013) Rapid mutation of Spirulina platensis by a new mutagenesis system of atmospheric and room temperature plasmas (ARTP) and generation of a mutant library with diverse phenotypes. PLOS ONE 8:e77046
Lee JC, Straffon MJ, Jang TY, Higgins VJ, Grant CM, Dawes IW (2001) The essential and ancillary role of glutathione in Saccharomyces cerevisiae analysed using a grande gsh1disruptant strain. FEMS Yeast Res 1:57–65
Toroser D, Yarian CS, Orr WC, Sohal RS (2006) Mechanisms of γ-glutamylcysteine ligase regulation. Biochim Biophys Acta 1760:233–244
Park HY, Nam MH, Lee HS, Sun HY, Jun WJ, Hendrich S, Lee KWW (2010) Isolation of caffeic acid from Perilla frutescensand its role in enhancingc-glutamylcysteine synthetase activity and glutathione level. Food Chem 119:724–730
Jozefczak MJ, Remans T, Vangronsveld J, Cuypers (2012) A glutathione is a key player in metal-induced oxidative stress defenses. Int J Mol Sci 13:3145–3175
Ramzan M, Mehmood T (2009) Enhanced production of glucose oxidase from UV-mutant of Aspergillus niger. Afr J Biotechnol 8:288–290
Routledge MN, Mirsky FJ, Wink DA, Keefer LK, Dipple A (1994) Nitrite-induced mutations in a forward mutation assay: influence of nitrite concentration and pH. Mutat Res 322:341–346
Nakamura S, Suzui N, Nagasaka T, Komatus F, Ishioka NS, Ito-Tanabata S, Kawachi N, Rai H, Hattori H, Chino M, Fujimaki S (2013) Application of glutathione to roots selectively inhibits cadmium transport from roots to shoots in oilseed rape. J Exp Bot 64:1073–1081
Gales G, Penninckx M, Block JC, Leroy P (2008) Role of glutathione metabolism status in the definition of somecellular parameters and oxidative stress tolerance of Saccharomyces cerevisiae cells growing as biofilms. FEMS Yeast Res 8:667–667
Toroser D, Sohal RS (2005) Kinetic characteristics of native γ-glutamylcysteine ligase in the aging housefly, Musca domestica L. Biochem Biophys Res Commun 326:586–593
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual. 3rd edn. Clod Spring Harbor Laboratory, New York
Stadler LJ (1928) Genetic effects of X-rays in maize. Proc Natl Acad Sci USA 14:69–75
Brash DE (2015) Invited review UV signature mutations. Photochem Photobiol 91:15–26
Ohtake Y, Yabuuchi S (1991) Molecular cloning of the γ-glutamylcysteine synthetase gene of Saccharomyces cerevisiae. Yeast 7:953–961
Biterova EI, Barycki JJ (2009) Mechanistic details of glutathione biosynthesis revealed by crystal structures of Saccharomyces cerevisiae glutamate cysteine ligase. J Biol Chem 28:32700–32708
Prajapati RS, Das M, Sreeramulu S, Sirajuddin M, Srinivasan S, K rishnamurthy V, Ranjiani R, Ramakrishhan C, Varadarajan R (2007) Thermodynamic effects of proline introduction on protein stability. Proteins 66:480–491
Gogos A, Shapiro L (2002) Large conformational changes in the catalytic cycle of glutathione synthase. Structure 10:1669–1676
Bajaj K, Madhusudhan MS, Adkar BV, Chakrabarti P, Ramakrishnan C, Sali A, Varadarajan R (2007) Stereochemical criteria for prediction of the effects of proline mutations on protein stability. PLoS Comput Biol 13:e241
Acknowledgements
The work was supported by the Science Research Plan of Hebei Higher Schools (No. Z2010225), open fund of Key laboratory (No. 3333112) and Bioengineering key discipline of Hebei Province.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Xu, W., Jia, H., Zhang, L. et al. Effects of GSH1 and GSH2 Gene Mutation on Glutathione Synthetases Activity of Saccharomyces cerevisiae . Protein J 36, 270–277 (2017). https://doi.org/10.1007/s10930-017-9731-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-017-9731-0