Abstract
Rice is the most important crop consumed all over the world. In Brazil, irrigated rice covers 50 % of the rice producing area and is responsible for 75 % of the national production. Upland rice covers most of the remaining area, and is therefore, a very important production system in the country. In the present study, we have used the drought tolerant upland rice variety Três Meses Antigo to investigate the proteomic changes that occur during drought stress. Plants were submitted to drought by the reposition of 50 % of the water lost daily. Twenty days after the beginning of the drought stress period, leaves were harvested and used for protein extraction. The 2D maps obtained from treated and control plants revealed 408 reproducible spots, 44 of which were identified by mass spectrometry, including 15 differential proteins. Several unaltered proteins were also identified (39 spots) and were mainly involved in photosynthesis. Taken together, the results obtained suggest that the tolerant upland rice up-regulates anti-oxidant and energy production related proteins in order to cope with water deficit.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Oryza sativa L. is one of the most cultivated cereals around the world and approximately three billion people depend on this culture daily for their nutrition. Rice is a staple food in both developing and developed countries and is cultivated by several different methods. In Brazil, the main production system in which rice is grown is the irrigated system, which occupies 75 % of the national production. However, production costs of irrigated rice are elevated, since high amounts of water and intensive soil preparation during the plant life cycle are required. Irrigated rice is also responsible for significant rates of methane emissions [1]. These factors impose restrictions to irrigated rice expansion. On the other hand, upland rice cultivation, which is another rice production system in Brazil, presents relatively reduced costs, low environmental impact and is an important activity for the small family farms and crop rotation of soybean and corn. Moreover, it is extremely important for the agriculture-pasture system, especially in the recovery of degraded pastures. However, the frequent occurrence of drought during the cultivation period increases the economic risks associated to this production system. In order to overcome this problem, breeding programs have been working on the development of cultivars with high yield, improved grain quality and tolerance to drought, especially considering recent concerns about water shortage and uneven rainfall distribution.
Rice was the first crop to have its genome deciphered (International Rice Genome Sequencing Project, 2005), and has been considered a reference in crop studies. Since the release of the rice genome sequence, several studies have been performed to understand gene function by using functional genomic tools [2–6]. Proteomics is one of these approaches, which has become a very important field of study for the comprehension of complex cellular processes since it represents the link between genomic information and protein composition present in a specific tissue. Indeed, an increasing amount of studies have been reported during the last 10 years using gel-based and gel-free Proteomic techniques. The results obtained reveal that although great progress has been achieved in MS-based techniques, 2-DE still accounts for 75 % of rice proteomic studies, indicating that these are complementary methodologies [7]. Regarding the field of study in rice proteomics, almost half of the proteomic reports in the last years have focused on abiotic stresses, including drought [7]. Recently, Singh and Jwa [8] reported that the most common rice proteome responses to all abiotic stresses include photosynthesis, redox homeostasis, detoxification/antioxidation pathway, carbohydrate metabolism, and protein metabolism. Drought is one of the main environmental factors that severely affects rice yield and therefore has been given considerable attention [9–13] however, very few studies have focused on upland rice varieties [2, 14, 15].
Improvement of drought tolerance has been a challenge and the comprehension of plant response to drought using a proteomic approach may help fulfill this task. In the present study, the proteome of the upland rice variety Três Meses Antigo, considered tolerant to drought stress [16], was analyzed using bidimensional electrophoresis (2-DE) and mass spectrometry, in order to identify proteins associated to drought tolerance.
2 Materials and Methods
2.1 Plant Material and Drought Conditions
Different upland rice (O. sativa L. var. japonica) varieties were grown on PVC pipe columns (25 cm of diameter; 80 cm of height) using fertilized Oxisol (0.3927 m3), which is the most common type of soil found in Brazil, under greenhouse conditions [16]. The experimental design and watering treatments were performed as previously described [14]. Briefly, the control condition consisted of well-watered plants, which received 100 % reposition of the water lost daily, while the drought stress consisted of 50 % reposition of the water lost daily from anthesis on. Leaf samples from plants in the control and water-stressed conditions were randomly collected at 20 days after initiating the drought stress treatment, frozen in liquid nitrogen and maintained at −80 °C. At harvest, several yield components were evaluated, such as harvest index, spikelet sterility, among others and drought tolerance parameters were estimated based on calculations of drought severity, drought tolerance index and drought susceptibility index, as described previously [14, 17]. In a previous study [16], varieties were classified according to their response to drought stress and the variety Três Meses Antigo was considered tolerant to water deficit and was therefore selected for proteomic analyses.
2.2 Protein Extraction and Image Analysis
Leaves collected from each replicate were pooled together and used for protein extraction as described previously [14]. Protein quantification was performed using the Bradford Reagent (Invitrogen, USA). Isoelectric focusing was performed with 11-cm immobilized pH gradient (IPG) strips with a pH range of 4–7 in a Multiphor II electrophoresis system (GE). Approximately 220 μg of protein were used to rehydrated IPG strips with 2 % (v/v) CHAPS, 8 M urea, 7 mg dithiothreitol (DTT) and 2 % IPG buffer. SDS–PAGE was performed in 10 % gels as described by Laemmli [18] and protein spots were visualized after silver [19] or Coomassie blue staining. Three high quality gels, each representing one biological replicate, of both conditions were selected for protein spot analysis using the ImageMaster 2D Platinum v7.0 (GE Healthcare, UK) as previously described [14]. Automatically detected spots were manually checked and spot alignment was improved by manual spot detection and matching. Proteins were considered differential when protein abundances in control and drought stressed plants were significantly different after Student’s t test (α = 0.05).
2.3 Protein Identification by Mass Spectrometry
Protein spots were excised manually from Coomassie stained gels and digested using sequencing grade trypsin (Promega, Madison, WI) according to Shevchenko [20]. Aliquots of digested proteins (1.0 μL) were mixed with a saturated solution of α-cyano-4-hydroxycinnamic acid and applied to a MALDI target plate. Mass spectra were acquired using a MALDI-TOF/TOF Autoflex II spectrometer (Bruker Daltonics, Bremen, Germany) according to Rabello et al. [14]. Peak lists were generated using the FlexAnalysis 3.0 software (Bruker Daltonics) and protein identification was obtained using the MASCOT program (Matrix Science, UK) with the NCBInr protein database and Oryza sativa taxonomy, using the parameters previously described [14].
3 Results and Discussion
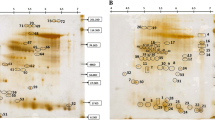
In this study we have analyzed an upland japonica rice variety (Três Meses Antigo), which showed tolerance to water deficit [16]. In a previous analysis, we compared the root tissue of two upland japonica rice genotypes (drought tolerant and susceptible) submitted to drought stress and observed that the susceptible plants responded to water deficit in a similar way as irrigated (indica) rice varieties. The tolerant variety showed an up-regulation of several proteins involved in cell protection in response to drought [14]. In the present study, we were interested in the protein profile changes of an upland drought tolerant variety submitted to water deficit when compared to well-watered conditions. Twenty days after the drought stress, leaves were sampled and used for protein extraction. The 2D maps revealed a total of approximately 408 proteins per gel varying in size from 10 to 220 kDa and in pI from 4 to 7 (Fig. 1). The comparison between the protein profiles of the control and the drought stressed plants revealed proteins responsive to drought (Table 1), as well as unaltered proteins (Table 2), some of which were identified by mass spectrometry in order to better understand the tolerance of the upland rice Três Meses Antigo.
3.1 Differentially Expressed Proteins
The comparison of the 2D maps of well-watered and drought-stressed plants was performed using silver-stained gels given the high sensitivity of this detection method. This analysis showed a total of 99 differentially expressed proteins, some of which were observed in Coomassie-stained gels and subjected to in gel digestion. A total of 15 differential protein spots were successfully identified by mass spectrometry (Table 1). Although several proteins presented significant differential intensity, as determined by the statistical analysis, a pronounced alteration in the protein profile in response to drought stress was not observed, as has been reported in irrigated rice studies [21]. In the present analysis, the differential spots identified (Fig. 2) showed fold differences varying between 1.2 and 2.4 (Table 1). Previous studies indicate that water deficit induces a series of responses in plants. These responses include: changes in protein translation involved in signaling; transcriptional control; protection of cellular components such as membranes and proteins; osmotic adjustment; and detoxification [22, 23]. The low levels of altered proteins observed in this study may suggest that the maintenance of the photosynthetic system as well as energy production is part of the tolerance process presented by the variety Três Meses Antigo. It is possible that after 20 days of water deficit, plants are adapted to the amount of available water and are able to maintain growth and production.
One of the differential proteins identified in the present study (3MA 26) was Rubisco activase, which was decreased in water stressed plants. Rubisco activase is a chloroplastic enzyme that regulates the activation of Rubisco [24], and is controlled indirectly by light intensity, which establishes a favorable H+ and Mg2+ gradient, and allows the synthesis of ATP, necessary for its activation [25].
Other proteins associated to photosynthetic metabolism were also identified, including spot 3MA 21, which was identified as putative precursor chloroplastic glutamine synthetase, and was decreased during water deficit. This enzyme is involved in the incorporation of ammonium into glutamine, which is the main precursor of several metabolites containing nitrogen. Therefore, glutamine synthetase may have an important role in plant survival during stress conditions. It has been reported that glutamine synthetase is much more active in plants under recovery of severe stress than under water deficit [26]. Since the sampling point for the proteome analysis was at 20 days of drought stress, it is possible that the plant had already adapted to the water deficit condition.
The general plant metabolism is also influenced by stress and proteins such as triosephosphatase isomerase (spot 3MA 66) and H protein subunit of glycine decarboxylase 3′ (spot 3MA 85) were differentially expressed. Spot 3MA 66 was increased in stressed plants while spot 3MA 85 was decreased.
Unexpectedly, the drought-induced S-like ribonuclease protein (spot 3MA 62) was decreased in the drought stress condition. According to Salekdeh [27], the abundance of this protein increases at the beginning of the drought stress (18 days after drought stress). After rewatering, plants showed a decrease in the abundance of this protein. Again, it is possible that, in our study, the plants were already adapted to the water deficit condition at the sampling time of 20 days after drought stress.
One protein involved in metabolism, identified in the present study as putative dihydrolipoamide dehydrogenase precursor (spot 3MA 16), was down-regulated in the drought condition (Table 1). This protein has been described as having a role in cell protection during saline stress in rice plantlets [28].
It has been reported that the up-regulation of proteins involved in cellular detoxication is a general defense mechanism in upland rice [14, 29]. Proteins that belong to this category, up-regulated during water deficit, were identified in this study, including putative germin-like protein (3MA 74) and ascorbate peroxidase (3MA 68). Berna [30] reported that the expression of germin-like protein is stimulated by biotic and abiotic stresses such as heavy metal stress and fungal infection. Peroxidases are antioxidant enzymes that have been described in roots of upland rice under water stress [29]. All these proteins have been reported as part of an essential response of tolerant plants to water deficit [29, 31, 32]. Although these proteins involved in protection against oxidative stress are primarily induced under water deficit, the plants under well-watered conditions also showed up-regulation of proteins related to this function. We identified a putative thioredoxin peroxidase (spot 3MA 84), which was decreased in well-watered plants.
Spot 3MA 31, identified in this study as a putative remorin 1 protein, increased in water-stressed plants and has been reported as induced in leaves of tobacco and Arabidopsis submitted to drought. This protein is associated to the membrane and therefore it is suggested that it plays a crucial role in drought tolerance [33, 34]. This protein has also been found in membrane fractions resistant to detergents [35]. Membranes have an important role in sensing stress and proteins associated to membranes can also play roles in the maintenance of cell turgor.
Overall, transcriptional studies in drought stressed plants suggest that several genes involved in photosynthesis are down-regulated during water deficit conditions [36–38]. In our analysis of the variety Três Meses Antigo submitted to drought we found a different response. Some photosynthesis proteins were down-regulated while proteins related to oxidative protection were up-regulated. The ability of regulating photosynthesis and oxidative stress mechanisms seem to be crucial for plant adaptation to drought stress. Taken together, the results seem to indicate that the variety used in this study does not show severe changes in the proteome profile in order to cope with water deficit. We sampled leaves several days after the beginning of drought stress since we were interested in evaluating the ability of the plant to cope with long periods of water deficit. It is possible that at 20 days after the beginning of the stress, the plant used mechanisms to adapt to the stressful condition without severely compromising its development and especially production. Indeed, Guimarães [16] observed that Três Meses Antigo revealed only 18 % reduction in grain yield under drought stress.
3.2 Unaltered Proteins Identified
Since the variety used in this study is tolerant to drought stress, we also identified some proteins which remained unaltered during the control and drought stress conditions (Table 2). A total of 39 out of 309 protein spots with unchanged expression were identified and were mostly involved in photosynthesis and energy production. Some proteins associated to protection against oxidative stress damage were also identified. It is possible that the maintenance of the expression of these proteins may play some role in the determination of drought tolerance. These proteins identified can be considered as part of the reference map of Três Meses Antigo and can be used for future comparisons with other varieties differing in their tolerance to drought.
Interestingly, an abscisic acid- and stress-inducible protein (spot 3MA 73) was identified but did not show a significant change in intensity under water deficit, as determined by the statistical test. The production and catabolism of ABA is a well known process and is delicately regulated by stress [39]. In water deficit conditions, ABA has a critical role regulating the water status in the plant through guard-cells, as well as by inducing the expression of genes that encode enzymes and other proteins involved in dehydration tolerance [40]. ABA production and regulation is highly important for cell survival, since it acts as a hormone that regulates stomatal function under stress conditions [41–45]. It is possible that due to the water deficit tolerance feature of Três Meses Antigo, this protein is expressed constitutively in order to protect the plant from a stress condition.
Several spots corresponding to Rubisco were identified, which did not show altered intensities in response to water deficit. Rubisco is one of the most affected proteins in susceptible plants, since photosynthetic activity is highly altered during drought stress [46]. Moreover, proteins associated to energy production such as putative mitochondrial Fo ATP synthase D chain (spot 3MA 81) and the oxygen-evolving complex protein 1 (spot 3MA 48), involved in photosystem II, were identified and are important for ATP production during photosynthesis. The up-regulation of oxygen-evolving complex protein was also observed by Ali [2] in rice leaf sheath during drought stress.
Other metabolism proteins were identified including a putative NAD-malate dehydrogenase (spot 3MA 42). Dooki [47] reported, through a proteomic approach, that the enzyme malate dehydrogenase was highly abundant in rice panicle subjected to abiotic stress. The up-regulation of this enzyme has also been observed under saline, water and cold stresses [48, 49]. The plastidic enzyme cysteine synthase (spot 3MA 30) was also identified and has been reported to be induced under cold stress in rice [50]. Cysteine synthase is related to sulfur metabolism and is intimately linked to the production of glutathione, a metabolite known to be involved in resistance to biotic and abiotic stresses [50]. Another protein involved in stress resistance identified was superoxide dismutase (spot 3MA 89). Antioxidant enzymes have been reported as part of an essential response of tolerant plants to water deficit [29, 31, 32].
4 Concluding Remarks
The comparison between the protein maps obtained from well-watered versus drought stressed upland rice plants allowed the identification of several differently accumulated proteins. These results contribute to a better understanding of the response of the tolerant genotype to water deficit. Proteins related to photosynthetic metabolism were abundantly expressed and some of them showed reduced levels upon water deficit. The results obtained suggest that the tolerant upland rice up-regulates anti-oxidant and energy production related proteins in order to cope with water deficit. Moreover, the capacity of maintaining protein expression mostly unaltered during water stress may also play a role in drought tolerance, especially proteins involved in photosynthesis. This study shows the capacity of proteomic studies for the identification of proteins potentially involved in plant tolerance to drought stress, a highly complex metabolic process. The understanding of this process is somewhat still limited, especially for upland varieties, which are naturally more tolerant to water deficit. Therefore, the identification of protein and genes of agronomic interest, allied to genetic improvement programs has been considered an important strategy for the development of varieties more adapted to water restriction conditions. Proteomics is currently considered an extremely valuable approach to reach this goal and can significantly contribute to rice breeding programs.
Abbreviations
- PVC:
-
Polyvinyl chloride
- ABA:
-
Abscisic acid
- 3MA:
-
Três meses antigo
- CHAPS-3:
-
[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DTT:
-
Dithiothreitol
- IPG:
-
Immobilized pH gradient
- SDS–PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- MALDI-TOF:
-
Matrix-assisted laser desorption/ionization-time-of-flight
- MS:
-
Mass spectrometry
- SNAP:
-
Sophisticated numerical annotation procedure
References
Aselmann I, Crutzen PJ (1989) Global distribution of natural freshwater wetlands and rice paddies, their net primary productivity, seasonality and possible methane emissions. J Atmos Chem 8:307–358
Ali GM, Komatsu S (2006) Proteomic analysis of rice leaf sheath during drought stress. J Proteome Res 5:396–403
Komatsu S (2005) Rice proteome database: a step toward functional analysis of the rice genome. Plant Mol Biol 59:179–190
Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Huang X, Han B (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20:1238–1249
Helmy M, Tomita M, Ishihama Y (2011) OryzaPG-DB: rice proteome database based on shotgun proteogenomics. BMC Plant Biol 11:63
Yang Y, Dai L, Xia H, Zhu K, Liu H, Chen K (2013) Protein profile of rice (Oryza sativa) seeds. Gen Mol Biol 36:87–92
Agrawal GK, Rakwal R (2011) Rice proteomics: a move toward expanded proteome coverage to comparative and functional proteomics uncovers the mysteries of rice and plant biology. Proteomics 11:1630–1649
Singh R, Jwa NS (2013) Understanding the responses of rice to environmental stress using proteomics. J Proteome Res 11:4652–4669
Mirzaei M, Soltani N, Sarhadi E, Pascovici D, Keighley T, Salekdeh GH, Haynes PA, Atwell BJ (2012) Shotgun proteomic analysis of long-distance drought signaling in rice roots. J Proteome Res 11:348–358
Kosova K, Vitamvas P, Prasil IT, Renaut J (2011) Plant proteome changes under abiotic stress—contribution of proteomics studies to understanding plant stress response. J Proteomics 74:1301–1322
Liu JX, Bennett J (2011) Reversible and irreversible drought-induced changes in the anther proteome of rice (Oryza sativa L.) genotypes IR64 and Moroberekan. Mol Plant 4:59–69
Mushtaq R, Katiyar S, Bennett J (2008) Proteomic analysis of drought stress-responsive proteins in rice endosperm affecting grain quality. J Crop Sci Biotech 4:227–232
Mirzaei M, Soltani N, Sarhadi E, Pascovici D, Keighley T, Salekdeh GH, Haynes PA, Atwell BJ (2012) Shotgun proteomic analysis of long-distance drought signaling in rice roots. J Proteome Res 1:348–358
Rabello AR, Guimaraes CM, Rangel PH, da Silva FR, Seixas D, de Souza E, Brasileiro AC, Spehar CR, Ferreira ME, Mehta A (2008) Identification of drought-responsive genes in roots of upland rice (Oryza sativa L). BMC Genom 9:485
Huang W, Bi T, Sun W (2010) Comparative analysis of panicle proteomes of two upland rice varieties upon hyper-osmotic stress. Front Biol 5:546–555
Guimarães C, Stone L, Rangel P, Ferreira M (2006) In: 2º Congresso Brasileiro da Cadeia Produtiva de Arroz/VIII. Resitência à seca: I Avaliação de genótipos de arroz de terras altas, com divergência genotípica, em condições controlada. Reunião Nacional de Pesquisa de Arroz
Fischer R, Maurer R (1978) Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust J Agric Res 29:897–912
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99
Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858
Ke Y, Han G, He H, Li J (2009) Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem Biophys Res Commun 379:133–138
Bray EA (1993) Molecular responses to water deficit. Plant Physiol 103:1035–1040
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Portis AR (1992) Regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase activity. Annu Rev Plant Physiol Plant Mol Biol 43:415–437
Campbell WJ, Ogren WL (1995) Rubisco activase activity in spinach leaf extracts. Plant Cell Physiol 36:215–220
Ferreira VM, Magalhães PC, Durães FOM, Oliveira LEMD, Purcino AÁC (2002) Metabolismo do nitrogênio associado à deficiência hídrica e sua recuperação em genótipos de milho. Ciência Rural 32:13–17
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2:1131–1145
Kumari S, Nee Sabharwal VP, Kushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A (2009) Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genomics 9:109–123
Wang H, Zhang H, Li Z (2007) Analysis of gene expression profile induced by water stress in upland rice (Oryza sativa L. var. IRAT109) seedlings using subtractive expressed sequence tags library. J Int Plant Biol 49:1455–1463
Berna A, Bernier F (1999) Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol Biol 39:539–549
Meloni DA, Oliva MA, Martinez CA, Cambraia J (2003) Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Eniron Exp Bot 49:69–76
Mittal R, Dubey RS (1991) Behaviour of peroxidases in rice: changes in enzyme activity and isoforms in relation to salt tolerance. Plant Physiol Biochem 29:31–40
Nelson CJ, Hegeman AD, Harms AC, Sussman MR (2006) A quantitative analysis of Arabidopsis plasma membrane using trypsin-catalyzed (18) O labeling. Mol Cell Proteomics 5:1382–1395
Valot B, Negroni L, Zivy M, Gianinazzi S, Dumas-Gaudot E (2006) A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteomics 6:145–155
Lefebvre B, Furt F, Hartmann MA, Michaelson LV, Carde JP, Sargueil-Boiron F, Rossignol M, Napier JA, Cullimore J, Bessoule JJ, Mongrand S (2007) Characterization of lipid rafts from medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol 144:402–418
Hazen SP, Pathan MS, Sanchez A, Baxter I, Dunn M, Estes B, Chang HS, Zhu T, Kreps JA, Nguyen HT (2005) Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array. Funct Integr Genomics 5:104–116
Wang D, Pan Y, Zhao X, Zhu L, Fu B, Li Z (2011) Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genom 12:149
Oztur ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48:551–573
Zhang J, Peng Y, Guo Z (2008) Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res 18:508–521
Zhu XF, Chen XW, Li XB, Qian Q, Huang DN, Zhu LH, Zhai WX (2002) Genetic mapping of T-DNA integration sites in Xa21 transgenic rice. Yi Chuan Xue Bao 29:880–886
Düaiucl H, Alleweldt G (1973) Der lahresgang der Abscisinsäure in vegetativen Organen von Reben. Vitis 12:26–32
Broquedis M (1987) L’acide abscissique et l’abscissate de B-D-glucopyranose dans le développement des baies de raisin, la germination des pépins et la formation des racines sur les boutures de Vigne. Vittis 28:121–135
Düring H, Bachmann O (1975) Abscisic acid analysis in Vitis vinifera in the period of endogenous bud dormancy by high pressure liquid chromatography. Physiol Plantarum 34:201–203
During H (1992) Gas exchange of grapcvines leaves as affected by soil factors. In: Simposium international sur la physiologie de la vign. In Comptes Rendas 295–298
Broquedis M, Bouard L (1989) L’acide abscissique dans différents aspects de la physiologie de la Vigne. Connaissanee de la Vigne et du Vin—Aspects Actuels de la Viticulture. In Hors Série 89–94
Tabaeizadeh Z (1998) Drought-induced responses in plant cells. Int Rev Cytol 182:193–247
Dooki AD, Mayer-Posner FJ, Askari H, Zaiee AA, Salekdeh GH (2006) Proteomic responses of rice young panicles to salinity. Proteomics 6:6498–6507
Imin N, Kerim T, Rolfe BG, Weinman JJ (2004) Effect of early cold stress on the maturation of rice anthers. Proteomics 4:1873–1882
Liska AJ, Shevchenko A, Pick U, Katz A (2004) Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant Physiol 136:2806–2817
Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics 5:484–496
Acknowledgments
This research was supported by Embrapa, CNPq and Embrapa Recursos Genéticos e Biotecnologia. The authors declare no competing financial interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rabello, F.R., Villeth, G.R.C., Rabello, A.R. et al. Proteomic Analysis of Upland Rice (Oryza sativa L.) Exposed to Intermittent Water Deficit. Protein J 33, 221–230 (2014). https://doi.org/10.1007/s10930-014-9554-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-014-9554-1