Abstract
Physiologically based pharmacokinetic (PBPK) models can be used to leverage physiological and in vitro data to predict monoclonal antibody (mAb) concentrations in serum and tissues. However, it is currently not known how consistent predictions of mAb disposition are across PBPK modelling platforms. In this work PBPK simulations of IgG, adalimumab and infliximab were compared between three platforms (Simcyp, PK-Sim, and GastroPlus). Accuracy of predicted serum and tissue concentrations was assessed using observed data collected from the literature. Physiological and mAb related input parameters were also compared and sensitivity analyses were carried out to evaluate model behavior when input values were altered. Differences in serum kinetics of IgG between platforms were minimal for a dose of 1 mg/kg, but became more noticeable at higher dosages (> 100 mg/kg) and when reference (healthy) physiological input values were altered. Predicted serum concentrations of both adalimumab and infliximab were comparable across platforms, but were noticeably higher than observed values. Tissue concentrations differed remarkably between the platforms, both for total- and interstitial fluid (ISF) concentrations. The accuracy of total tissue concentrations was within a three-fold of observed values for all tissues, except for brain tissue concentrations, which were overpredicted. Predictions of tissue ISF concentrations were less accurate and were best captured by GastroPlus. Overall, these simulations show that the different PBPK platforms generally predict similar mAb serum concentrations, but variable tissue concentrations. Caution is therefore warranted when PBPK models are used to simulate effect site tissue concentrations of mAbs without data to verify the predictions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pharmacological targets of monoclonal antibodies (mAbs) are often located outside the vasculature, which makes extracellular tissue concentrations a potentially more informative measure of pharmacodynamic (PD) effects than serum concentrations [1]. Tissue penetration of mAbs is typically poor, resulting in steady-state tissue-to-serum concentration ratios between 0.05 and 0.15 [2]. Due to their large size (~ 145 kDa), diffusion of mAbs through vascular endothelial cell membranes is slow and convective transport through paracellular pores can be considered as the main mechanism of uptake of mAbs to tissues [3]. Transcytosis mediated by the neonatal Fc receptor (FcRn) is an additional possible mechanism of extravasation, but its quantitative importance in mAb tissue disposition is not yet clear [4]. Additionally, the extent of mAb distribution to tissues is further hampered by tissue clearance through lymphatic drainage, endosomal catabolism and target-mediated drug disposition (TMDD) [5]. Tissue distribution of mAbs is also dependent on the type of tissue, with lower concentrations commonly observed in tissues with tightly knit vascular endothelial membranes, such as the brain, compared to tissues with more discontinuous capillaries [2]. Due to these differences between serum and effect-site concentrations, it becomes important to accurately characterize the extent and rate of antibody distribution to the tissue of interest.
Antibody tissue distribution can be characterized experimentally through biodistribution studies. These studies are costly however, and are almost exclusively done in preclinical animals due to their invasive nature [3]. An alternative and relatively new approach is to use microdialysis to assess tissue concentrations, which is less invasive and specifically measures interstitial fluid (ISF) concentrations [6]. Since most targets are located in the extracellular space, ISF concentrations can be considered to be more pharmacologically relevant than total tissue concentrations, which reflect a mean concentration lumped over extra- and intracellular tissue sub-compartments. However, microdialysis has up to now only sporadically been used for the assessment of mAb distribution [6, 7]. Given the cost and limitations of biodistribution studies, the development of supporting in silico models of mAb distribution might aid in understanding and predicting local effect site concentrations.
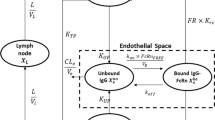
One way to predict mAb tissue and serum concentrations is through physiologically based pharmacokinetic (PBPK) modeling. PBPK models consist of sets of ordinary differential equations (ODEs) which link drug concentrations in different organs/tissues, represented as compartments. The models are parameterized using physiological data such as tissue volumes, lymph flows and drug related (in vitro) data, such as molecular weight and target affinity [8]. PBPK models were originally developed for small molecule drugs but they are, in adapted form, increasingly being used for therapeutic proteins such as mAbs. The first PBPK model for mAbs was developed by Covell and colleagues in 1986 and described IgG tissue concentrations in mice [9]. Their model was later expanded to include more mAb disposition mechanisms, such as mAb target binding, convective transport using a two pore formalism [10] and FcRn-mediated recycling [11]. In 2012, Shah and Betts developed a PBPK model of mAb disposition capable of predicting mAb tissue concentrations in humans and different preclinical species (mice, rats and cynomolgus monkeys) [12]. Similar models were developed and packaged by three different PBPK software platforms, namely Simcyp [13], PK-Sim [14] and GastroPlus [15]. While the underlying structure of the models is comparable (Fig. 1), some differences in input parameters (e.g. lymph flows) between the models have been implemented which was to be expected, given the number of values required and the variation in reported literature sources and estimation approaches observed. Thus far, it has not been assessed whether these three platforms provide comparable predictions.
Generalized representation of a tissue compartment in a physiologically based pharmacokinetic (PBPK) model. The intracellular space and competition of monoclonal antibody (mAb) and endogenous IgG for FcRn are not shown. \({\mathrm{f}}_{\mathrm{up}}^{\mathrm{vasc}}\): fraction taken up from serum, \({\mathrm{f}}_{\mathrm{rc}}^{\mathrm{vasc}}\): fraction recycled to serum, \({\mathrm{K}}_{\mathrm{d}}\): mAb-FcRn dissociation constant (either at pH = 7.4 or pH = 6.0), \({\mathrm{K}}_{\mathrm{e},\mathrm{cat}}\): catabolic elimination rate constant, \({\mathrm{K}}_{\mathrm{rc}}\): recycling rate constant,\({\mathrm{K}}_{\mathrm{up}}\): uptake rate constant, \(\mathrm{L}\): lymph flow, \(\mathrm{PS}\): permeability surface area product for large (\({\mathrm{PS}}_{\mathrm{L}}\)) or small (\({\mathrm{PS}}_{\mathrm{S}}\)) pores, \(\mathrm{Q}\): blood flow,\({\upsigma }_{\mathrm{i}}\):lymphatic reflection coefficient, \({\upsigma }_{\mathrm{v}}\): vascular reflection coefficient for large (\({\upsigma }_{\mathrm{v},\mathrm{L}}\)) or small (\({\upsigma }_{\mathrm{v},\mathrm{S}}\)) pores
The aim of this work was therefore to compare serum and tissue predictions of mAbs using the three most commonly used PBPK platforms (Simcyp, PK-Sim and GastroPlus). First, the behavior of a generic mAb with the properties of an IgG was evaluated using serum and tissue concentrations (total, ISF and endosomal concentrations). Next, differences in physiological input parameters were compared across the platforms. To evaluate the behavior of the models under altered circumstances, the influence of changing key physiological and mAb-related parameters on serum and tissue (skin ISF) exposure was compared across the platforms. To investigate the accuracy of the predictions of mAb serum kinetics, model predictions were compared with published observed data originating from five different clinical studies investigating the PK of adalimumab [16, 17] and infliximab [18,19,20] at different intravenous doses. Lastly, the accuracy of tissue disposition predictions was evaluated using reported antibody distribution coefficients (ABC, i.e. the tissue/serum concentration ratios).
Methods
Physiologically based pharmacokinetic models
PBPK simulations of monoclonal antibody disposition were done using three different PBPK platforms; Simcyp (v20, Certara, Inc, NJ, USA), PK-Sim® (v11, Open Systems Pharmacology) and GastroPlus (v9.8.2, Simulations PlusInc., Lancaster, California, USA). A generalized model structure is presented graphically in Fig. 1. In short, each tissue compartment is divided into a vascular-, endosomal-, interstitial- and intracellular space. mAbs are delivered from the arterial blood supply to the vascular tissue space using the arterial tissue blood flow (\(Q\)) and recirculate to the venous blood supply using the venous tissue blood flow, which is the arterial tissue blood flow minus the lymph flow (\(L\)) (\(Q-L\)). mAbs can translocate to the interstitial space through convective transport via paracellular (endothelial) pores and/or transcytosis mediated via FcRn. The magnitude of these trans-capillary transport processes depends on the convective lymph flow (\(L\)), the vascular reflection coefficient (\({\sigma }_{v}\)) and the permeability surface-area product (PS). From the vasculature and/or interstitial space, mAbs are taken up in the endothelial endosomes through pinocytosis (rate constant \({K}_{up}\)). Inside these endosomes, mAbs interact with FcRn receptors according to a 1:1 stoichiometry using a mAb-specific dissociation constant (\({K}_{d,pH6}\)). This mAb binding to FcRn happens in competition with endogenous IgG. Bound mAb either recycles to the vascular- or interstitial space (rate constant\({K}_{rc}\)). Unbound mAb is degraded by the acidic endosomal environment using a first order rate constant (\({K}_{e,cat}\)). The relative proportion of uptake from- and recycling to the vascular space (relative to the interstitial space) is controlled by the constants \({f}_{vasc,up}\) and \({f}_{vasc,rc}\), respectively. From the interstitial space, mAbs are transported to the lymph nodes using the lymph flow (\(L\)) and the lymphatic reflection coefficient (\({\sigma }_{l})\). From the lymph nodes the mAbs are transported to the venous serum supply using the lymph flow (\(L\)). The intracellular space is considered inaccessible for mAbs, except through target mediated drug disposition (TMDD) in case of membrane-bound antigens.
Simulation settings
Physiological population input values required for the PBPK models were parameterized using the pre-specified reference populations in the platforms (“Healthy volunteer” for Simcyp, “European” for PK-Sim and “American” for GastroPlus). From each of these populations, one 24-year old male individual was sampled with a fixed body weight of 80.7 kg and height of 176.6 cm. This resulting virtual reference individual was used in all subsequent simulations.
Simulations were done using three different compounds; a generic mAb (IgGexo), and two TNF-a inhibitors (adalimumab and infliximab). The compound specific input parameters are presented in Table 1. For IgGexo, the compound input values specified in the simulator for endogenous IgG were used without adaptation, meaning that the substrate behaves as an endogenous IgG as specified in the platforms. For the TNF-a inhibitors, identical input parameters were used across platforms. As TMDD for soluble antigens cannot currently be specified in GastroPlus and requires interfacing with another program in PK-Sim (MoBi, Open Systems Pharmacology), simulations for the TNF-a inhibitors were initially done without TMDD. To evaluate the effect of TMDD on model predictions, a second set of predictions was made with the Simcyp platform with TMDD enabled (“Simcyp + TMDD”). TNF-a abundance and degradation rate were set at 4.47 pM and 0.877/h, respectively (standard values in the simulator).
Quantitative comparison of model outputs
Simulated concentration–time profiles of the mAbs in serum and tissues were used as primary outputs. Only tissue compartments with a similar implementation across platforms were compared (details: supplementary Table S1). Gastro-intestinal-tract concentrations for example were not compared as this tissue is implemented as “gut” in Simcyp, divided into separate compartments (“stomach”, “small intestine” and “large intestine”) in PK-Sim and is not available for outputting in GastroPlus. Both total- and interstitial fluid (ISF) concentrations were evaluated. In PK-Sim, total tissue concentrations were assessed as “whole organ” concentrations with a residual blood fraction of 0.18 for all tissues except brain. This fraction is recommended to be used when comparing simulations with concentrations obtained through tissue dissection studies [14] (i.e. the verification data used in this work). For total brain concentrations, a residual blood fraction of 0 is used, as the verification data for brain stemmed from a study where concentrations were corrected for residual blood content [25]. The R-package “PKNCA” [26] was used to calculate non-compartmental PK parameters. Tmax was defined as the time when the maximum concentration (Cmax) is observed. As output concentrations were sampled at 4 points per hour, Tmax estimates had a precision of 15 min. The AUC was either calculated to the last observed timepoint (AUC0-last, e.g. AUC0–672 h) or by extrapolating the last observed concentration (\({C}_{last})\) to infinity (AUCinf), using the estimated elimination rate constant (\({\lambda }_{z})\). Other parameters were calculated as follows: (Eqs. 1–4)
with CL: clearance,\({V}_{d}\): volume of distribution, \({t}_{1/2}\): half-life and ABC: antibody distribution coefficient.
Parameters were quantitatively compared between platforms using the percent coefficient of variation (%CV). Values larger than 50% were specified as denoting a large disagreement between models. Agreement between model outcomes and observed (literature) data was expressed as fold errors for individual observations and absolute average fold errors (AAFEs) for multiple (n) observations; Eqs. (5–6).
with x: the parameter of interest. FEs and AAFEs within twofold (0.5–2.0) were deemed acceptable. When observed data had to be extracted from figures, Webplotdigitizer [27] was used.
Results
Behavior of a generic mAb (IgG exo )
To investigate differences between the platforms in simulating PK of exogenous IgG (IgGexo), serum and tissue concentration–time profiles of a mAb with the same input parameters as those defined for endogenous IgG were simulated using the different platforms (Table 1). Limited differences in serum kinetics were observed for a dose of 1 mg/kg of IgGexo (Fig. 2). For this dose, serum AUC0-672h values were comparable across the three platforms (5787, 6570, 5571 mg h/L for Simcyp, PK-Sim and GastroPlus respectively, 9% CV), as were the half-lives (570, 635 and 614h for Simcyp, PK-Sim and GastroPlus respectively, 5% CV) (Supplementary Table S2). However, the associated ISF and total tissue concentration profiles differed significantly between the platforms (Fig. 3). Variation in AUC0-672h based on total tissue concentrations was less than 50% CV for adipose (42% CV), heart (41% CV), kidney (35% CV) and muscle tissue (6% CV) (Supplementary Table S2). For AUC0–672 h values calculated from ISF concentrations, only heart and lung values varied less than 50% CV (12 and 42% CV, respectively). When tissue concentrations were normalized for serum concentration, i.e. representing antibody distribution coefficients (ABC), differences between platforms remained large for ISF and total tissue concentrations for most tissues. The rate of distribution also differed between platforms, with time to maximum tissue concentrations (Tmax) varying significantly for most tissues (6/9 and 5/9 of tissues for total and ISF concentrations, respectively) (Supplementary Table S2). Endosomal concentrations also varied across platforms (Supplementary Figure S1). In PK-Sim and GastroPlus, endosomal concentrations were comparable across tissues, whereas in Simcyp, the concentrations depended more on the specific tissue (e.g. low in adipose but high in heart tissue). Dosages of 0.1 and 10 mg/kg yielded near identical dose-normalized results in serum compared to the 1 mg/kg dose (Supplementary Figure S2). Dosages of 100 mg/kg and higher showed decreased dose-normalized exposure (AUC0–84 days), but these decreases differed markedly between platforms. For a 1000 mg/kg dose for example, decreases in dose-normalized exposure were − 33%, − 54% and − 3.2% relative to a 1 mg/kg dose for Simcyp, PK-Sim and GastroPlus, respectively (Supplementary Table S3).
Predicted tissue concentrations of exogenous IgG (IgGexo) after a single 1mg/kg intravenous dose using three different physiologically based pharmacokinetic (PBPK) platforms; Simcyp (full blue line), PK-Sim (dashed red line) and GastroPlus (dot-dashed green line). Shown concentrations are total tissue concentrations (A) and interstitial fluid tissue concentrations (B) (Color figure online)
Comparison of model parameters
Next, differences in model parameter values across platforms were investigated, together with model behavior when key parameters were altered. First, physiological input parameters were compared. These parameter values are presented in Fig. 4 and associated %CV between platforms are given in supplementary Table S4. In a next step, the effect of changing key input parameter values on the AUC0–672 h in serum and the relative penetration in the interstitial space of the skin (AUC0–672 h ratio skin ISF /serum) was investigated. Skin was used as an example tissue for this analysis due to its relevance for inflammatory disease states (e.g. psoriasis). The results of the analysis are presented in Fig. 5, together with associated serum and skin ISF concentration profiles in supplementary Figure S3.
Sensitivity analyses on the impact of changing input parameters on serum exposure (A) and relative interstitial fluid (ISF) exposure in skin tissue (B). Results are expressed as percentage changes from the baseline area under the curve (AUClast) value of 1mg/kg exogenous IgG (IgGexo). Initial parameter values were either halved (left hand plots) or doubled (right hand plots). For visualization purposes, the + 281% change in relative skin exposure when large pore size was doubled in PK-Sim was capped to 75% (denoted with an asterix (*)). CO: cardiac output, FcRn: endosomal FcRn receptor abundance, IgG: endogenous IgG serum concentration, Kd (pH 6): FcRn-mAb dissociation constant at endosomal pH, MWT: molecular weight of the mAb, R,s (skin): pore size of small pores in skin tissue, R,l (skin): pore size of large pores in skin tissue
Tissue volumes
Total tissue volumes were comparable across the platforms (minimum 4% CV (heart); maximum 33% CV (lung)). The relative fractions of intracellular volume were also similar across platforms (minimum 3% CV (adipose); maximum 44% CV (lung)), as were the relative interstitial tissue volumes, except for the brain (104% CV, other tissues maximum: 48% CV in the heart). In contrast, relative endosomal tissue volumes varied considerably between platforms (maximum: 154% CV in adipose tissue) due to the high values included in GastroPlus. The relative vascular volume of the tissues also varied considerably across platforms, with only three tissues having a % CV under 50% being skin (0% CV), muscle (35% CV) and brain (43% CV).
Blood and lymph flows
Blood flows were comparable, with an observed maximum of 30% CV (adipose tissue) between platforms. Lymph flows varied more between platforms, with only 2 of the 9 tissues having %CVs under 50% (muscle and skin). When cardiac output was altered, blood and lymph flows scaled proportionally across all platforms, as blood flows are specified as percentages of cardiac output and lymph flows as a fraction of those blood flows. Halving or doubling the cardiac output led to similar minor decreases and increases in serum exposure across platforms, respectively. The relative disposition into the ISF of the skin followed an inverse pattern in PK-Sim and GastroPlus (i.e. higher relative exposure when blood flow is decreased), while in Simcyp, the relative ISF skin disposition was almost unaltered with changes in blood flow.
Endosomal uptake and recycling
Endosomal uptake rate constants and recycling rate constants varied considerably between platforms (166 and 111% CV, respectively) due to the high values implemented in PK-Sim and Simcyp, respectively. The fractional uptake from the vasculature and the fraction recycled to the vasculature control the direction of endosomal uptake and recycling processes. In PK-Sim, there is no uptake from- and recycling to the interstitial space as both parameters are set to 1. In GastroPlus, there is uptake from and recycling to the interstitial space but this is minor compared to uptake from and recycling to the vasculature (parameters set to 0.72). In Simcyp, there is equal endosomal uptake from the vasculature and interstitial space (fractional uptake from vasculature: 0.5), but the direction of recycling depends on the tissue (to the interstitial space for adipose, muscle and skin; to the vasculature for the other tissues).
Endosomal catabolism
The catabolic elimination rate constant and endosomal FcRn concentration varied noticeably between platforms (> 162% CV and > 68% CV, respectively) due to the respectively high and low parameter values specified in GastroPlus.
When endosomal FcRn abundance was halved, serum exposure decreased in all three models although the magnitude of this decrease varied markedly (between − 12% and − 42% decrease of AUC0–672 h in GastroPlus and Simcyp, respectively). Doubling the FcRn abundance had an opposite effect on serum exposure as AUC0–672 h increased between 6 (GastroPlus) and 26% (Simcyp). Relative skin ISF exposure only changed as a result of alterations in FcRn abundance in the Simcyp platform: relative skin ISF exposure was lower or higher when FcRn abundance was decreased or increased, respectively. Similar, but smaller changes in serum and skin ISF exposure were induced when the affinity of the mAb (IgGexo) for FcRn was doubled or halved. Changes in endogenous IgG serum concentration only had a noticeable impact in the Simcyp model, where lower endogenous IgG serum concentrations yielded higher serum and relative skin ISF exposure of the mAb (IgGexo), probably due to less competition for FcRn receptors. Changing the Kd of IgGexo had a similar impact on serum exposure across the platforms. The impact of changing the Kd on relative skin exposure was however only noticeable in the Simcyp model.
Trans-capillary transport
Trans-capillary transport between the vasculature and the interstitial space is implemented in Simcyp and PK-Sim through convective and passive transport through small and large pores. The magnitude of these processes depends on the pore sizes of the membrane and the solute size of the mAb. mAb solute sizes are calculated from the molecular weight of the mAb using two slightly different formulas (Supplementary Figure S4a). For a mAb with a molecular weight of 150 kDa for example, the estimated solute size was 5.08 nm in Simcyp and 5.13 nm in PK-Sim. Similarly, the reflection coefficients determining the magnitude of convection are calculated from mAb solute size and pore size using different formulas (Supplementary Figure S4b). For a mAb with a solute size of 5 nm, the calculated reflection coefficient for a pore size of 20 nm was 0.31 in Simcyp and 0.23 in PK-Sim. When the solute size is fixed to a default value of 5.34 nm instead of estimated, the differences in concentration–time profiles of the mAb in serum (Supplementary Figure S4c) and ISF skin (Supplementary Figure S4d) are minor compared to existing between-platform differences. Further details on the calculation methods of convective and passive trans-capillary transport can be found in the supplementary detailed results section. In GastroPlus, only the convection process is implemented for which no distinction is made between small and large pore transport and reflection coefficients are empirically specified (standard value of 0.95).
When the molecular weight of the mAb was halved, the serum exposure decreased in Simcyp (− 15%) and PK-Sim (− 14%) and the relative skin ISF exposure increased (+ 54.1% and + 65.5%, respectively). When mAb molecular weight was doubled, the impact was reversed. Doubling or halving the molecular weight of the mAb had no impact on the serum or skin ISF disposition in GastroPlus. Halving or doubling the pore sizes of endothelial skin pores had different impacts on serum and relative skin ISF disposition depending on the platform (PK-Sim or Simcyp) and whether the pores were large or small.
Accuracy of model predictions
Accuracy of serum predictions of adalimumab and infliximab
To investigate the accuracy of mAb serum kinetics prediction, model outputs were compared with published observed data originating from five different clinical studies investigating the PK of adalimumab [16, 17] and infliximab [18,19,20] at different intravenous doses. The simulated profiles are presented in supplementary Figure S5 (adalimumab) and Figure S6 (infliximab) and show clear overpredictions during the elimination phase. The overall accuracy of the serum profiles of adalimumab, expressed as the absolute average fold error (AAFE) was similar across platforms; 1.36 for Simcyp, 1.40 for PK-Sim and 1.55 for GastroPlus. When TMDD was incorporated in the Simcyp model, the AAFE slightly decreased to 1.34. For infliximab, the AAFEs were higher: 1.95 for Simcyp, 1.96 for PK-Sim and 2.28 for GastroPlus. Incorporation of TMDD in the Simcyp model only led to a minor decrease in AAFE to 1.94. Calculated NCA parameters for the 5 mg/kg dose were similar across platforms for both mAbs (%CV < 25%) and addition of TMDD to the Simcyp model only marginally decreased the half-life (− 0.8%) (Table 2). Predicted AUCinf, t1/2 and Vd were higher than observed values, while CL was underestimated in the simulations.
Accuracy of antibody distribution coefficients
Next, the accuracy of tissue predictions was assessed by comparing published and PBPK-predicted ABCs. Table 3 gives the observed and predicted ABCs for the different tissues, together with fold-errors. ABCs for total concentrations (ABCtot) were compared with the values aggregated by Shah and Betts [2] (all tissues except brain) and data from a dedicated study in brain tissue [25]. Predicted ABCtot were within a twofold of the observed values, except for adipose (underpredicted in PK-Sim), brain (overpredictions in Simcyp and GastroPlus, underpredicted in PK-Sim), heart (underpredicted in PK-Sim), lung (overpredicted in PK-Sim) and skin (underpredicted in GastroPlus and PK-Sim) tissue. The overall AAFE of ABCtot across tissues, with the exception of brain, was 1.22 for Simcyp, 1.81 for PK-Sim and 1.39 for GastroPlus. Predicted ABCs for interstitial fluid tissue concentrations (ABCISF) were compared with the average ABCISF values originating from four studies [7, 28,29,30] (values and study descriptions in supplementary Table S5). Predicted ABCISF were less accurate with none of the tissues being consistently predicted within a factor of two across all platforms. The largest errors were noted for liver (> threefold overpredicted in all platforms), brain (> tenfold underpredicted in Simcyp, > threefold overpredicted in PK-Sim) and spleen (> threefold overpredicted in PK-Sim). The overall AAFE of ABCISF for all tissues except brain was 2.50 for Simcyp, 2.30 for PK-Sim and 1.96 for GastroPlus.
Discussion
PBPK models can aid in predicting mAb kinetics in serum and clinically inaccessible tissues [8]. There are several software platforms available which enable PBPK modelling of mAbs through a graphical user interface. These platforms offer increased reusability over custom tailored models, but their comparative predictive performance has yet to be evaluated. Therefore, it is uncertain whether the choice of modelling platform noticeably impacts predictions made. To address this gap, three major PBPK modelling platforms (Simcyp, PK-Sim and GastroPlus) were compared in terms of model behavior and accuracy of serum and tissue predictions of intravenously administered mAbs.
The PBPK models compared in this study generally predicted similar mAb serum concentrations, and concentration–time profiles show a bi-exponential decline indicative of redistribution of mAbs from tissues that quickly equilibrate with serum to less permeable tissues. One can note slightly lower initial concentrations using the GastroPlus model and overall higher concentrations in the PK-Sim model. Although these differences are minor and have little impact on the total exposure (AUC), they might be indicative of larger differences at the tissue level.
The models diverged more in terms of serum predictions at high dosages or when physiological input parameters, originating from a healthy reference population, were altered. At high dosages, saturation of the endosomal FcRn receptors, which protect the mAb from endosomal degradation, can result in elevated catabolism of the mAb [31]. The simulations carried out in this work indicate that this non-linear behavior is incorporated in all three models but that the saturation of FcRn receptors starts to occur at different dosages depending on the platform. More specifically, profiles simulated in GastroPlus remain linear with doses up to 1000 mg/kg, while at that dose, normalized exposure has already dropped substantially in Simcyp and PK-Sim. While these dosages are much higher than conventional mAb dosages, they are in the range of high dose intravenous immunoglobulin (IVIG) therapy [32, 33]. Therefore, care should be taken when using the models to predict serum concentrations of IgG after IVIG therapy. At the more conventional doses (1 mg/kg), the platforms also diverge in serum predictions when physiological parameters are changed. For example, changing endogenous IgG concentration by twofold has a clear effect on serum PK in Simcyp but the impact is negligible in GastroPlus and PK-Sim. Altering cardiac output or FcRn abundance on the other hand show similar directional changes with some differences in magnitude between platforms. This is important, as these discrepancies might indicate that disease effects (e.g. lower/higher endogenous IgG levels) might have differing impacts on predicted PK depending on the platform.
When key drug-related input parameters such as Kd and molecular weight were changed, the models exhibited expected behavior, with decreased serum exposure of mAbs when their affinity for FcRn is lower (i.e. high Kd, leading to more catabolism) or when mAb molecular weight is decreased (i.e. smaller size, resulting in more extravasation to tissues) [28, 34]. One limitation of these analyses is that the initial parameter values were platform-specific. As a result, the initial parameter value was in many cases not harmonized across platforms and differences in response to altered parameters might also be partially due to these different initial values if non-linearities are present. This approach was chosen over an approach with harmonized initial parameter values to better mimic a typical PBPK modelling workflow, where initial physiological parameter values are not altered a priori.
To assess the accuracy of serum disposition predictions for a common set of drug-specific input parameters, predictions of adalimumab and infliximab were compared with observed data collected from the literature. The behavior of these mAbs in serum was almost identical across platforms but the elimination phase was clearly miss-specified compared to observed data (t1/2 overpredicted). Addition of TMDD did not resolve this overprediction. With the data at hand, it is not possible to know whether these inaccuracies are due to mis-specified PBPK models or a biased in vitro Kd value used as input. A parameter estimation procedure can typically be used in a PBPK modeling workflow to estimate an alternative Kd value that better fits observed data. However, this was not done here since the goal was to assess the accuracy of the models in absence of observed clinical data. One limitation of the accuracy assessment was that subcutaneous (SC) dosing was not assessed, despite being a common route of administration for mAbs. This was on purpose since not all platforms had a prespecified module to mechanistically predict mAb absorption after SC dosing.
Although the serum predictions were fairly accurate and generally comparable across platforms, this was mostly not the case for the tissue predictions. The accuracy of the projected tissue concentrations was low and highly dependent on the specific tissue and the applied PBPK platform. For total tissue concentrations, the accuracy in brain tissue was particularly poor, as two platforms predicted concentrations which were more than ten times higher than observed values (Simcyp and GastroPlus). Wu and colleagues have shown with their mice brain PBPK model that more anatomically relevant models are needed to adequately capture brain tissue concentrations [35]. It should also be noted that the observed data for brain concentrations were corrected for residual blood content (using 51Cr-labeled red blood cells) [25], but that the total tissue concentration outputs of the platforms also encompass the vascular compartment. PK-Sim has the option to alter the fraction of residual blood available for sampling, and this fraction was set to zero for brain concentrations, which might explain why more accurate results are obtained for total brain concentrations in this platform. For observed total concentrations of other tissues, no formal correction for residual blood content was applied on the experimental (tissue dissection) data. However, some studies used by Shah and Betts to estimate aggregated ABC values applied a whole body perfusion procedure with phosphate buffered saline (PBS) prior to sampling, aimed at the removal of residual blood from the tissues, potentially biasing the reference data towards lower concentrations [2].
Overall, the predictive performance was higher for total concentrations than for ISF concentrations. GastroPlus had the better performance in terms of ISF concentrations than Simcyp and PK-Sim, as only two tissues had inaccurate ABCs (i.e. outside twofold) compared to six in Simcyp and PK-Sim. This is important for target engagement assessments for example, as pharmacological tissue targets are typically located in the interstitial space, either as soluble or membrane- bound targets [1]. Again, the variability in the observed data should also be highlighted, as ISF concentrations estimated from observed total concentrations (assuming no intracellular penetration) [28, 30] and from microdialysis studies [7, 29] were aggregated (supplementary Table S5) and ABCISF values varied considerably between studies for a given tissue and method (supplementary Figure S7).
One limitation to also consider in this accuracy assessment is that the observed ABC values were primarily based on tissue concentrations in rodents, since tissue concentrations in humans are scarce. In their analysis of ABC values collected from the literature, Shah and Betts noted that the ABC values for a given tissue are comparable across species, making pooling of values appropriate [2]. Furthermore, Mandikian and colleagues compared tissue physiology across different preclinical species and humans and found that tissue volumes (vascular and interstitial) and blood flows scale well allometrically [36]. In contrast, the affinity of the FcRn-IgG interaction appears to be different in mice than in humans [37]. Additionally, due to a lack of tissue concentration–time profiles obtained in human tissues, no statements can be made regarding the accuracy of the initial shape (i.e. distribution phase) of the simulated profiles.
The discrepancies observed in tissue predictions between platforms were expected given the inter-platform differences in physiological input parameters and model assumptions. For instance, as mAbs are “cleared” from the interstitial space by the lymphatic system, the different values specified in the platforms might be partially responsible for differences in ISF concentrations. Due to a lack of clinical data, parameters values such as lymph flows are often estimated or assumed. The most notable difference in model assumptions between platforms is the way FcRn mediated transcytosis is implemented. Currently, the role and quantitative relevance of transcytosis in mAb tissue distribution has not been fully elucidated [3]. In vitro experiments have confirmed bidirectional FcRn mediated transcytosis of mAbs [38, 39], but a reduction in relative tissue exposure in FcRn knockout-mice has only been observed in a few tissues (muscle and skin) [4]. Thus, more in vitro and clinical data are needed to better inform the parameter values and underlying assumptions, especially for endothelial pore sizes, lymph flows, and rate constants (uptake-, recycling- and catabolic elimination rate constants).
An additional limitation of this work is that we were unable to identify an ideal set of assumptions and physiological input parameter values which optimize the accuracy of tissue concentrations. The reason for this was because there were some gaps in the available documentation across platforms, mainly relating to the exact mathematical model structure (i.e. ODEs) and a few intermediary calculations (e.g. passive diffusion, see supplementary results). These gaps precluded reproducing the models in an independent platform (e.g. R). Such a translation step would be required to uniformly detect influential variables (e.g. through a global sensitivity analysis) and to estimate appropriate input variable values conditional on the observed data. While sensitivity analyses and parameter estimation tools exist in all platforms, they differ in scope and methodology, which makes comparing results across platforms difficult. This is why a crude but uniform twofold change in input parameters was applied as sensitivity analysis in the current work.
Conclusion
Overall, the PBPK platforms performed similarly when serum concentrations of mAbs were simulated at standard dosages under normal physiological conditions. Tissue concentrations and serum mAb concentrations simulated at higher dosages or under pathophysiological circumstances were more variable across platforms. This study illustrates the need for additional physiological data and experimental insights in mAbs’ disposition. When using PBPK to assess effect-site concentrations in tissues, the model verification procedure should not be confined to serum only but also include an accuracy assessment on the tissue(s) of interest.
References
Tang Y, Li X, Cao Y (2021) Which factors matter the most? Revisiting and dissecting antibody therapeutic doses. Drug Discovery Today 26:1980–1990. https://doi.org/10.1016/j.drudis.2021.04.022
Shah DK, Betts AM (2013) Antibody biodistribution coefficients. MAbs 5:297–305. https://doi.org/10.4161/mabs.23684
Glassman PM, Abuqayyas L, Balthasar JP (2015) Assessments of antibody biodistribution. J Clin Pharmacol 55:S29–S38. https://doi.org/10.1002/jcph.365
Garg A, Balthasar JP (2007) Physiologically-based pharmacokinetic (PBPK) model to predict IgG tissue kinetics in wild-type and FcRn-knockout mice. J Pharmacokinet Pharmacodyn 34:687–709. https://doi.org/10.1007/s10928-007-9065-1
Lobo ED, Hansen RJ, Balthasar JP (2004) Antibody pharmacokinetics and pharmacodynamics. J Pharm Sci 93:2645–2668. https://doi.org/10.1002/jps.20178
Jadhav SB, Khaowroongrueng V, Derendorf H (2016) Microdialysis of large molecules. J Pharm Sci 105:3233–3242. https://doi.org/10.1016/j.xphs.2016.08.016
Dragatin C, Polus F, Bodenlenz M et al (2016) Secukinumab distributes into dermal interstitial fluid of psoriasis patients as demonstrated by open flow microperfusion. Exp Dermatol 25:157–159. https://doi.org/10.1111/exd.12863
Liu S, Shah DK (2022) Mathematical models to characterize the absorption, distribution, metabolism, and excretion of protein therapeutics. Drug Metab Dispos 50:867–878. https://doi.org/10.1124/dmd.121.000460
Covell DG, Barbet J, Holton OD et al (1986) Pharmacokinetics of monoclonal immunoglobulin G1, F(ab’)2, and Fab’ in mice. Cancer Res 46:3969–3978
Baxter LT, Zhu H, Mackensen DG, Jain RK (1994) Physiologically based pharmacokinetic model for specific and nonspecific monoclonal antibodies and fragments in normal tissues and human tumor xenografts in nude mice. Cancer Res 54:1517–1528
Ferl GZ, Wu AM, DiStefano JJ (2005) A predictive model of therapeutic monoclonal antibody dynamics and regulation by the neonatal Fc receptor (FcRn). Ann Biomed Eng 33:1640–1652. https://doi.org/10.1007/s10439-005-7410-3
Shah DK, Betts AM (2012) Towards a platform PBPK model to characterize the plasma and tissue disposition of monoclonal antibodies in preclinical species and human. J Pharmacokinet Pharmacodyn 39:67–86. https://doi.org/10.1007/s10928-011-9232-2
Gill KL, Gardner I, Li L, Jamei M (2016) A bottom-up whole-body physiologically based pharmacokinetic model to mechanistically predict tissue distribution and the rate of subcutaneous absorption of therapeutic proteins. AAPS J 18:156–170. https://doi.org/10.1208/s12248-015-9819-4
Niederalt C, Kuepfer L, Solodenko J et al (2018) A generic whole body physiologically based pharmacokinetic model for therapeutic proteins in PK-Sim. J Pharmacokinet Pharmacodyn 45:235–257. https://doi.org/10.1007/s10928-017-9559-4
Zhou H, Bolger M, Lukacova V (2015) Application of PBPK modeling to predict monoclonal antibody disposition after intravenous and subcutaneous administration in rats and humans [ABSTRACT]. In: AAPS. Orlando, FL
den Broeder A, van de Putte L, Rau R et al (2002) A single dose, placebo controlled study of the fully human anti-tumor necrosis factor-alpha antibody adalimumab (D2E7) in patients with rheumatoid arthritis. J Rheumatol 29:2288–2298
Weisman MH, Moreland LW, Furst DE et al (2003) Efficacy, pharmacokinetic, and safety assessment of adalimumab, a fully human anti-tumor necrosis factor-alpha monoclonal antibody, in adults with rheumatoid arthritis receiving concomitant methotrexate: a pilot study. Clin Ther 25:1700–1721. https://doi.org/10.1016/S0149-2918(03)80164-9
Palaparthy R, Udata C, Hua SY et al (2018) A randomized study comparing the pharmacokinetics of the potential biosimilar PF-06438179/GP1111 with Remicade® (infliximab) in healthy subjects (REFLECTIONS B537–01). Expert Rev Clin Immunol 14:329–336. https://doi.org/10.1080/1744666X.2018.1446829
Park W, Lee SJ, Yun J, Yoo DH (2015) Comparison of the pharmacokinetics and safety of three formulations of infliximab (CT-P13, EU-approved reference infliximab and the US-licensed reference infliximab) in healthy subjects: a randomized, double-blind, three-arm, parallel-group, single-dose, Phase I study. Expert Rev Clin Immunol 11:25–31. https://doi.org/10.1586/1744666X.2015.1090311
Kavanaugh A, St Clair EW, McCune WJ et al (2000) Chimeric anti-tumor necrosis factor-alpha monoclonal antibody treatment of patients with rheumatoid arthritis receiving methotrexate therapy. J Rheumatol 27:841–850
EMEA (2004) Humira EPAR - scientific disussion
FDA (1999) Remicade (infliximab) for IV injection - supplement approval
Suzuki T, Ishii-Watabe A, Tada M et al (2010) Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. J Immunol 184:1968–1976. https://doi.org/10.4049/jimmunol.0903296
Kaymakcalan Z, Sakorafas P, Bose S et al (2009) Comparisons of affinities, avidities, and complement activation of adalimumab, infliximab, and etanercept in binding to soluble and membrane tumor necrosis factor. Clin Immunol 131:308–316. https://doi.org/10.1016/j.clim.2009.01.002
Garg A, Balthasar JP (2009) Investigation of the influence of FcRn on the distribution of IgG to the brain. AAPS J 11:553–557. https://doi.org/10.1208/s12248-009-9129-9
Denney WS, Duvvuri S, Buckeridge C (2015) Simple, automatic noncompartmental analysis: the PKNCA R package. J Pharmacokinet Pharmacodyn 42(11–107):S65. https://doi.org/10.1007/s10928-015-9432-2
Rohatgi A (2021) Webplotdigitizer: Version 4.5
Rafidi H, Rajan S, Urban K et al (2022) Effect of molecular size on interstitial pharmacokinetics and tissue catabolism of antibodies. mAbs 14:2085535. https://doi.org/10.1080/19420862.2022.2085535
Jadhav SB, Khaowroongrueng V, Fueth M et al (2017) Tissue distribution of a therapeutic monoclonal antibody determined by large pore microdialysis. J Pharm Sci 106:2853–2859. https://doi.org/10.1016/j.xphs.2017.03.033
Chang HP, Cheung YK, Shah DK (2021) Whole-body pharmacokinetics and physiologically based pharmacokinetic model for monomethyl Auristatin E (MMAE). J Clin Med 10:1332. https://doi.org/10.3390/jcm10061332
Jin F, Tayab ZR, Balthasar JP (2006) Pharmacokinetic and pharmacodynamic effects of high-dose monoclonal antibody therapy in a rat model of immune thrombocytopenia. AAPS J 7:E895-902. https://doi.org/10.1208/aapsj070487
Viala M, Vinches M, Alexandre M et al (2018) Strategies for clinical development of monoclonal antibodies beyond first-in-human trials: tested doses and rationale for dose selection. Br J Cancer 118:679–697. https://doi.org/10.1038/bjc.2017.473
Mahmood I, Tegenge MA, Golding B (2020) Considerations for optimizing dosing of immunoglobulins based on pharmacokinetic evidence. Antibodies 9:24. https://doi.org/10.3390/antib9020024
Ryman JT, Meibohm B (2017) pharmacokinetics of monoclonal antibodies. CPT: pharmacometrics & systems. Pharmacology 6:576–588. https://doi.org/10.1002/psp4.12224
Wu S, Le Prieult F, Phipps CJ et al (2022) PBPK model for antibody disposition in mouse brain: validation using large-pore microdialysis data. J Pharmacokinet Pharmacodyn 49:579–592. https://doi.org/10.1007/s10928-022-09823-x
Mandikian D, Figueroa I, Oldendorp A et al (2018) Tissue physiology of cynomolgus monkeys: cross-species comparison and implications for translational pharmacology. AAPS J 20:107. https://doi.org/10.1208/s12248-018-0264-z
Abdiche YN, Yeung YA, Chaparro-Riggers J et al (2015) The neonatal Fc receptor (FcRn) binds independently to both sites of the IgG homodimer with identical affinity. MAbs 7:331–343. https://doi.org/10.1080/19420862.2015.1008353
Dickinson BL, Badizadegan K, Wu Z et al (1999) Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J Clin Invest 104:903–911. https://doi.org/10.1172/JCI6968
McCarthy KM, Yoong Y, Simister NE (2000) Bidirectional transcytosis of IgG by the rat neonatal Fc receptor expressed in a rat kidney cell line: a system to study protein transport across epithelia. J Cell Sci 113:1277–1285. https://doi.org/10.1242/jcs.113.7.1277
Funding
This work received no external funding.
Author information
Authors and Affiliations
Contributions
Conceptualization: AV, EG, P-JDS; Methodology: AV, EG, P-JDS; Formal analysis and investigation: P-JDS; Writing—original draft preparation: P-JDS; Writing—review and editing: AV, EG, P-JDS; Supervision: AV.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
De Sutter, PJ., Gasthuys, E. & Vermeulen, A. Comparison of monoclonal antibody disposition predictions using different physiologically based pharmacokinetic modelling platforms. J Pharmacokinet Pharmacodyn (2023). https://doi.org/10.1007/s10928-023-09894-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10928-023-09894-4