Abstract
Obesity is a chronic disease with an increasing impact on the world’s population. In this work, we present a method of identifying obesity automatically using text mining techniques and information related to body weight measures and obesity comorbidities. We used a dataset of 3015 de-identified medical records that contain labels for two classification problems. The first classification problem distinguishes between obesity, overweight, normal weight, and underweight. The second classification problem differentiates between obesity types: super obesity, morbid obesity, severe obesity and moderate obesity. We used a Bag of Words approach to represent the records together with unigram and bigram representations of the features. We implemented two approaches: a hierarchical method and a nonhierarchical one. We used Support Vector Machine and Naïve Bayes together with ten-fold cross validation to evaluate and compare performances. Our results indicate that the hierarchical approach does not work as well as the nonhierarchical one. In general, our results show that Support Vector Machine obtains better performances than Naïve Bayes for both classification problems. We also observed that bigram representation improves performance compared with unigram representation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Obesity is a chronic disease that has become a major public health issue [1]. According to the World Health Organization, in 2014, 1.9 billion adults were overweight, out of which 600 million were obese [2]. In Chile, according to the National Health Survey of 2010, 25 % of the adult population is affected by this condition [1]. Obesity is a disease that is often accompanied by health risks called comorbidities. These comorbidities can affect various systems of the body leading to complications related to insulin resistance, high blood pressure, high cholesterol, risks of coronary heart disease, ischemic stroke and type 2 diabetes mellitus among others [3–5].
Today it is not rare to see patient information being stored in electronic health records (EHR). The use of EHR has enabled researchers to develop information extraction systems to obtain information about different health risks and conditions that may affect patients [6].
Since obesity has become a major global change, there is a growing interest in studying this disease, including its related comorbidities. There have been some attempts to develop applications to improve knowledge, diagnosis, treatment and follow-up of obese patients [4, 7–12]. As an example, we can cite the work by Bordowitz et al. [13] in which the authors investigated whether implementing automatic calculation of body mass index (BMI) improved clinical documentation and obesity treatment. Regarding extraction of obesity and its comorbidities, we should mention the challenge to create information extraction systems to automatically identify and extract information about obesity and its comorbidities organized by the Informatics for Integrating Biology & the Bedside (i2b2) in 2008 [14]. They released a set of de-identified medical discharge records from Partners HealthCare Research Patient Data Repository. The records were annotated by two obesity experts who identified and assigned labels to obesity and each of its fifteen most frequent comorbidities.Footnote 1 The labels were assigned according to the textual documented information or intuitive judgment. Yang et al. [15] and Solt et al. [16] obtained the best results in this challenge both in textual extraction and intuitive judgment. Yang et al. [15] used a set of lexical and semantic resources, such as concepts, sub-concepts, synonyms, treatments and related symptoms, most of them from the Unified Medical Language System (UMLS). The resultant features were exploited by dictionary look-up, rule-based and machine learning methods. For the textual task, they obtained a macro-averaged F-measure of 81 % and for the intuitive task a macro-averaged F-measure of 63 %. On the other hand, Solt et al. [16] used a context-aware rule-based semantic classifier. To perform a semantic analysis of the records, they included a set of clue terms for each disease, such as synonyms, frequent typos and abbreviations among others. In the textual task they obtained a macro- averaged F-measure of 80 % and in the intuitive tasks a macro-averaged F-measure of 67 %.
The work of Murtaugh et al. in [17] describe a more recent approach to automatically extracted information related to obesity. They developed a Regular Expression Discovery Extractor (REDEx) to extract body weight-related measures, such as weight, height, abdominal circumference and BMI from clinical notes. They obtained an accuracy of 98.3 %, and an F measure of 98.5 %.
In this article, we present a method to identify obesity automatically, using text mining techniques and information related to body weight measures and obesity comorbidities from Electronic Medical Records (EMR) in Spanish. As our dataset, we used outpatient reports obtained from Guillermo Grant Benavente Hospital (HGGB). We proposed two classification approaches: a hierarchical and non-hierarchical one. Our work will face two main challenges: to identify obesity based on its comorbidities and other associated information and to process medical records in Spanish.
Materials and methods
Dataset description
We used as our dataset a total of 66,179 outpatient records obtained from the HGGB EMR system. Among the records, we have 46 medical specialties that registered information between 2011 and 2012. Each medical record has structured and non-structured fields. The structured fields make it possible to report risk factors (type 2 diabetes mellitus, hypertension, cardiovascular risk, among others), habits (sedentary lifestyle, smoking, alcoholism and drug use status), and vital signs (arterial pressure, blood sugar, cholesterol levels, among others). The non-structured or narrative fields make it possible to report physical examinations, medical history, observations, and indications. Some of the structured fields also included a small space for the doctor to register comments and observations relevant to the field. For the purpose of this work, we considered both narratives and structured fields.
Preprocessing
This stage had four main steps. First, we normalized each report.Footnote 2 Second, we replaced all the BMI values present in the text to its minimum value, according to its category (see Table 1).
Third, we created a customized dictionary of comorbidities associated with obesity. As our base list, we used the fifteen diseases provided in [14] plus two diseases provided by the annotators: Cushing disease and hypothyroidism. We expanded this list to create our customized dictionary by adding all the linguistic and clinical variants of each of the comorbidities. At the end of this process, we had a dictionary containing 507 tokens.
Finally, we used a custom-made dictionary of keywords related to obesity, body weight measures and/or BMI to clean our dataset by filtering out records that did not contain terms present in the dictionary. At the end of the preprocessing stage, we recovered a total of 3105 records containing information relevant to the study.
Annotation
We defined two classifications problems. In the first there were: obesity (O), overweight (OW), normal weight (NW), and underweight (UW). The second classification problem included the types in the obesity category: super obesity (S), morbid obesity (M), moderate obesity (MO), and severe obesity (SO) [18, 19].
To generate a gold standard for classification, we asked two students with a biomedical background to revise and annotate a total of 3105 records using an annotation tool designed in QT-designerFootnote 3 and programmed in Python. For each record, they first assigned a label within the first classification problem. If an annotator assigned O to a record, s/he was asked to annotate the record with a label from the second problem classification. We also asked the annotators to provide information about keywords related to obesity, body weight or obesity comorbidities present in the records but not considered in the list of keywords.
When the annotators finished labeling all the documents, we filtered out documents that reviewers deemed to be possible false positives. These were documents that mentioned keywords related to obesity but were not relevant to the study (e.g. “lower molecular weight”). At the end we asked a third annotator to solve any disagreement and also to validate the assigned classes.
Finally, we obtained a total of 3015 annotated documents for the first classification problem and 1180 annotated records for the second problem. We evaluated inter-annotator agreement using Cohen’s kappa coefficient [20]. Cohen’s kappa is a statistical index to measure agreement between two raters. When an inter-annotator agreement is poor, values closer to zero are expected. On the other hand, when the agreement is almost perfect, values between 0.81–1 are expected.
For the first classification problem (classes O, OW, NW, and UW), we obtained a k = 0.97. For the second classification problem (classes S, M, SO, and MO), we obtained a k = 0.96. This result indicates that there is almost perfect agreement between the annotators with regard to both problems [20]. Thus, our gold standard can be considered reliable and useful to build models and evaluate classification results.
Feature extraction for classification
To extract features for classification, first we filtered out records that did not contain information related to obesity comorbidities. The filtering process used a dictionary of obesity comorbidities that contains 17 diseases and regular expressions. After this process, we obtained a total of 2428 records. Second, we tokenized the resultant records using unigrams (N1) and bigrams (N2). We defined unigrams as single word tokens and bigrams as sequences of two-word tokens. We obtained a total of 2904 unigram tokens and 5834 bigram tokens. Before using these tokens as features for classification, we applied feature selection using the InfoGainAttributeEval filter together with Ranker [21] available from Weka.Footnote 4 The InfoGainAttributeEval filter selects features by measuring the information gain for the class. The Ranker method sorts the features by their individual evaluations scores obtained in the InfoGainAttributeEval. Using both methods we reduced our feature set to 500 in the first classification problem for both unigram and bigram tokens. For the second classification problem, we have 532 unigrams and 548 bigrams. These features contain the selected 500 features plus some tokens related to obesity types, such as “BMI 20”, “obesity degree”, and “severe obesity”, that we manually added to the set.
Feature representation
For each classification problem, we used Bag of Words (BoW) representation [22]. We used term frequency-inverse document frequency (TF-IDF) weighting schema to represent the occurrences of the selected features in each record [23]. Equations (1) and (2) describe the TF-IDF schema where TF is the term frequency, IDF is the inverse document frequency, D is the number of documents in the collection, {d in D: t in d} documents where the term t appears
Classification and evaluation
In this stage, we decided to build two classification approaches: one treating each of the classification problems separately and the other one simulating a hierarchical classification for the second problem under the O class. Both problems used as classifiers the implementations of Naïve Bayes (NB) and Support Vector Machine (SVM) provided in the library scikit for machine learning in Python [24]. NB classifiers are a family of probabilistic classifiers; the method assumes that features in the dataset are mutually independent [25]. In this problem, we use an implementation of NB with a multinomial approach together with TF-IDF matrix representation. SVM are supervised learning models that build a set of hyperplanes in a high dimensional space that can separate the classes and find the one that maximizes the margin between the members of the classes [25]. In the case of SVM, we used a linear kernel together with the one by one multiclass classification setting, and kept the rest of the parameters at their default values.
To evaluate the classification models, we implemented ten-fold cross validation and repeated each experiment 10 times in order to get a reliable error estimate [26, 27]. Performance measures used to evaluate the classifiers’ predictive capacity were Accuracy (ACC), F-measure, False Positive Rate (FPR) and False Negative Rate (FNR). We averaged all the performance measures over the ten runs. Equations (3) to (6) show how we calculated each performance measures, where TP: true positives, TN: true negatives, FP: false positives, and FN: false negatives. To compare performances, we calculated the weighted average of each performance measure using as weights the number of examples per class and used a paired t-test (significance level of 0.05).
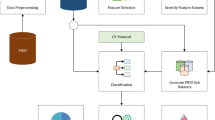
As mentioned earlier, we also implemented a small simulation of a hierarchical classification. Figure 1 explains the algorithm we used. We believed this implementation would only affect the second classification problem. The difference between the hierarchical method and the nonhierarchical one is given in the evaluation stage (lines 22–28), where we only considered TP examples from the O class to be part of the test set of the second problem. Examples with a label different from O do not have available the set of features to distinguish between obesity types. Furthermore, only a small fraction of examples labeled with O have such information (see lines 15 and 24).
Results
Annotation process results
Table 2 describes the dataset distribution after the annotation process. This table indicates to us that a class imbalance is affecting both classification problems.
Regarding gender, 80.84 % of the annotated records correspond to women having some degree of obesity. Another important result is that 93.19 % of the patients reported to be sedentary were also reported as suffering from obesity.
In Fig. 2, we observe that only 4.56 % of 66,179 retrieved records have information related to the presence or absence of obesity. Within that 4.56 %, only 39.13 % of the records mentioned information about the obesity type. We can also observe that the narrative fields were more informative regarding the presence or absence of the disease.
Figure 3 shows the distribution of the medical records with information related to this study among the different medical specialties.
Figure 4 shows the distribution of the main comorbidities among patients with and without obesity. We observe that hypertension and diabetes mellitus are the ones with the highest prevalence among obese patients. Although we have a list of seventeen comorbidities, Fig. 4 only shows comorbidities with more than 1 % of prevalence.
Classification results
Tables 3 and 4 show the classification results for both classification problems.
We can observe from Table 3 that both unigram and bigram representations together with SVM perform better than NB in terms of ACC and F-measure. We also observe that the O class obtains the highest values of FPR. On the other hand, the classes OW and NW have the highest values of FNR. Regarding weighted average, we observe that SVM with both unigram and bigram representations performs better than NB. From Table 4 we can observe that both unigram and bigram representations together with SVM, perform better than NB regarding single and weighted ACC and F-measure.
We also observe from Table 4 that the M class obtains the highest values of FPR for both unigram and bigram representations. On the other hand, the class S has high values of FNR. In general, from Table 3 and 4, we observe that N2 representation obtains better performance values.
When we implemented our hierarchical algorithm, we observed that only the second classification problem was affected in terms of performance. The results we obtained for the first classification problem were the same than those shown in Table 3. From Table 5, we observe that the performance obtained by our hierarchical method is lower than the one obtained in Table 4. In general, SVM performs better than NB.
Discussion and conclusion
This work shows a method to extract obesity from clinical records in Spanish by studying the disease, its comorbidities, body weight measures and BMI. The records do not have any explicit negation for the condition of obesity. Thus, we had to add counterexamples based on the nutritional information of the patient. Only 4.56 % of 66,179 available records have information related to this study.
According to the annotated records, women have the highest prevalence of obesity with an 80.84 % of the total of reported cases. We think the highest prevalence of obesity in women might be because women tend to visit health centers more frequently than men.
We used two approaches to treat the problem of classifying obesity and obesity types. The first approach treated the problem as two multiclass independent classification problems. The second approach used a hierarchical algorithm proposed by us, where the first classification problem was the first level of the hierarchy and the second classification problem was the second level of the hierarchy, with the O class as the parent class. Results showed that the nonhierarchical approach performed, in general, better than the hierarchical one. For the second classification level we only considered TP as candidate examples to be added in the test set of the second level; then the classification error from the first level of the hierarchy was, in a way, propagated to the second level of the hierarchy.
For both approaches, we observed the highest percentages of ACC. We explain this result by the high amount of TN obtained by both classification problems. We believe this result is due to the class imbalance observed for both classification problems (see Table 2). For this reason, we calculated a weighted average for Accuracy and F-measure. The weighted average showed, in general, lower ACC values when compared with single ACC values.
In most of the cases, SVM outperforms NB for both classification problems. In general, N2 representation shows better performance values than N1 representation. We believe that using N2 representation helps to capture more informative features (e.g. blood pressure, Gastroesophageal reflux disease, Type I, BMI 40). However, the computational cost of programming N2 is highest than programming N1 to extract features.
It is worth mentioning that the comorbidities are not exclusive of obesity, which could have generated ambiguities in the classifiers’ learning. In the first classification problem, the system tends to more often classify examples in class O, which generates a high percentage of FPR. We believed this affected the detection of examples in the NW and OW classes that present a high FNR. We can observe something similar in the M class, which has the highest percentages of FPR while the S-class shows a high FNR, except for NB of the hierarchical method. For the second classification problem, we observe that for both classifiers, N2 shows lowest FNR, except for the S class, when compared with N1 except for the S class in the hierarchical method with SVM. We have observed ambiguities in the use of the S class in the medical records. Sometimes the physician identifies a patient as having morbid obesity when it should be a super-obese patient. We believe that if we merge the S class with the M class, our classification results may improve.
Classifiers’ performance depends heavily on the selected features. Applying feature selection, in general, improved the performance of the classifiers when compared with classifiers built without feature selection. For this reason, in this work we decided to report the results obtained with feature selection.
Although the hierarchical approach showed itself to be slightly worse than the nonhierarchical one, the hierarchical approach is more realistic if we plan in future research to implement obesity, obesity types and comorbidities extraction as part of an EMR system in real time. The extraction system will give clinicians valuable information that will allow further studies related to obesity, its causes and related diseases.
Notes
Asthma, atherosclerotic cardiovascular disease, congestive heart failure, depression, diabetes mellitus, gallstones/cholecystectomy, gastroesophageal reflux disease, gout, hypercholesterolemia, hypertension, hypertriglyceridemia, obstructive sleep apnea, osteoarthritis, peripheral vascular disease, and venous insufficiency.
Words were changed to lower case and non-alphanumeric characters and stop words (e.g., a, the, on, etc.) were removed.
QT-designer is a Qt tool to design and build graphical user using widgets. http://doc.qt.io/qt-5/qtdesigner-manual.html
Weka is an open source software for data mining tasks. http://www.cs.waikato.ac.nz/ml/weka/
References
Atalah, E., Epidemiología de la obesidad en Chile. Revista Médica Clínica las Condes. 23(2):117–123, 2012.
Curtis, M., The obesity epidemic in the Pacific Islands. Journal of Development and Social Transformation. 1:37–42, 2004.
Markowitz, S., Friedman, M. A., and Arent, S. M., Understanding the relation between obesity and depression: causal mechanisms and implications for treatment. Clin. Psychol. Sci. Pract. 15(1):1–20, 2008.
Ergün, U., The classification of obesity disease in logistic regression and neural network methods. J. Med. Syst.. 33(1):67–72, 2009.
Guh, D. P., Zhang, W., Bansback, N., Amarsi, Z., Birmingham, C. L., and Anis, A. H., The incidence of co-morbidities related to obesity and overweight: a systematic review and meta-analysis. BMC Publ. Health. 9-88, 2009.
Crawford, A. G., Cote, C., Couto, J., Daskiran, M., Gunnarsson, C., Haas, K., Haas, S., Nigam, S. C., and Schuette, R., Prevalence of obesity, type II diabetes mellitus, hyperlipidemia, and hypertension in the United States: findings from the GE Centricity Electronic Medical Record database. Popul. Health Manag. 13(3):151–161, 2010.
Wood, G. C., Chu, X., Manney, C., Strodel, W., Petrick, A., Gabrielsen, J., Seiler, J., Carey, D., Argyropoulos, G., Benotti, P., Still, C. D., and Gerhard, G. S., An electronic health record-enabled obesity database. BMC Med. Inform. Decis. Mak. 12(1):1–8, 2012.
Ayash, C. R., Simon, S. R., Marshall, R., Kasper, J., Chomitz, V., Hacker, K., Kleinman, K. P., and Taveras, E. M., Evaluating the impact of point- of-care decision support tools in improving diagnosis of obese children in primary care. Obesity. 21(3):576–582, 2013.
Smith, A. J., Skow, A., Bodurtha, J., and Kinra, S., Health information technology in screening and treatment of child obesity: a systematic review. Pediatric. 131(3):e894–e902, 2013.
Cochran, J., and Baus, A., Developing interventions for overweight and obese children using electronic health records data. On-line Journal Of Nursing Informatics. 19(1):1–9, 2015.
Heydari, S. T., Ayatollahi, S. M., and Zare, N., Comparison of artificial neural networks with logistic regression for detection of obesity. J. Med. Syst. 36(4):2449–2454, 2012.
Kuebler, M., Yom-Tov, E., Pelleg, D., Puhl, R., and Muennig, P., When overweight is the normal weight: an examination of obesity using a social media internet database. PLoS ONE. 8(9):1–8, 2013.
Bordowitz, R., Morland, K., and Reich, D., The use of an electronic medical record to improve documentation and treatment of obesity. Fam. Med. 39(4):274–279, 2007.
Uzuner, Ö., Recognizing obesity and comorbidities in sparse data. J. Am. Med. Inform. Assoc. 16(4):561–570, 2009.
Yang, H., Spasic, I., Keane, J. A., and Nenadic, G., A text mining approach to the prediction of disease status from clinical discharge summaries. J. Am. Med. Inform. Assoc. 16(4):596–600, 2009.
Solt, I., Tikk, D., Gál, V., and Kardkovács, Z. T., Semantic classification of diseases in discharge summaries using a context-aware rule-based classifier. J. Am. Med. Inform. Assoc. 16(4):580–584, 2009.
Murtaugh, M. A., Gibson, B. S., Redd, D., and Zeng-Treitler, Q., Regular expression-based learning to extract bodyweight values from clinical notes. J. Biomed. Inform. 54:186–190, 2015.
NIH, NOEI, NHLBI, NAASO, The practical guide identification, evaluation, and treatment of overweight and obesity in adults, NIH Publication Number 0O-4084, 2000
Date, R. S., Walton, S. J., Ryan, N., Rahman, S. N., and Henley, N. C., Is selection bias toward super obese patients in the rationing of metabolic surgery justified?—A pilot study from the United Kingdom. Surg. Obes. Relat. Dis. 9(6):981–986, 2013.
Viera, A. J., and Garrett, J. M., Understanding interobserver agreement: the kappa statistic. Fam. Med. 37(5):360–363, 2005.
Amrita, M., Performance analysis of different feature selection methods in intrusion detection. International Journal of Scientific & Technology Research. 2(6):225–231, 2013.
Joachims, T., Learning to classify text using support vector machines. Vol. 1. New York: Engineering and Computer Sciences, 2002.
Gebrekidan, B., Zampieri, M., Wittenburg, P. T. H., Improving native language with TF-IDF weighing, Eighth Workshop on Innovative Use of NLP for Building Educational Applications. Atlanta, Georgia, 2013.
Buitinck, L., Louppe, G., Blondel, M., Pedregosa, F., Mueller, A., API design for machine learning software: experiences from the scikit-learn project, Paper presented at the European Conference on Machine Learning and Principles and Practices of Knowledge Discovery in Databases, 2013.
Rennie, J. D. M., and Rifkin, R., Improving multiclass text classification with the support vector machine. Cambridge: Massachusetts Institute Oftechnology, MIT, 2001.
Vanwinckelen, G., Blockeel, H., On estimating model accuracy with repeated cross-validation, 21st Belgian-Dutch Conference on Machine Learning, 2012.
Witten, I. H., Frank, E., Hall M. A., Data mining: practical machine learning tools and techniques, Third Edition, Series in Data Management Systems, Morgan Kaufmann, 2011.
Acknowledgments
The authors want to thank the informatics division of the Hospital Guillermo Grant Benavente (HGGB), Universidad de Concepcion, and Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT Grant N°11121463) for their support for this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Patient Facing Systems
Rights and permissions
About this article
Cite this article
Figueroa, R.L., Flores, C.A. Extracting Information from Electronic Medical Records to Identify the Obesity Status of a Patient Based on Comorbidities and Bodyweight Measures. J Med Syst 40, 191 (2016). https://doi.org/10.1007/s10916-016-0548-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-016-0548-8