Abstract
Electrocardiogram (ECG) compression finds wide application in various patient monitoring purposes. Quality control in ECG compression ensures reconstruction quality and its clinical acceptance for diagnostic decision making. In this paper, a quality aware compression method of single lead ECG is described using principal component analysis (PCA). After pre-processing, beat extraction and PCA decomposition, two independent quality criteria, namely, bit rate control (BRC) or error control (EC) criteria were set to select optimal principal components, eigenvectors and their quantization level to achieve desired bit rate or error measure. The selected principal components and eigenvectors were finally compressed using a modified delta and Huffman encoder. The algorithms were validated with 32 sets of MIT Arrhythmia data and 60 normal and 30 sets of diagnostic ECG data from PTB Diagnostic ECG data ptbdb, all at 1 kHz sampling. For BRC with a CR threshold of 40, an average Compression Ratio (CR), percentage root mean squared difference normalized (PRDN) and maximum absolute error (MAE) of 50.74, 16.22 and 0.243 mV respectively were obtained. For EC with an upper limit of 5 % PRDN and 0.1 mV MAE, the average CR, PRDN and MAE of 9.48, 4.13 and 0.049 mV respectively were obtained. For mitdb data 117, the reconstruction quality could be preserved up to CR of 68.96 by extending the BRC threshold. The proposed method yields better results than recently published works on quality controlled ECG compression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Electrocardiography (ECG) is the primary diagnostic tool for investigation of cardiac diseases. The amplitudes and time durations of the constituent waves P, QRS and T and wave segments PQ, ST and TP represent diagnostic information on cardiac functions. With the introduction of microcomputers in medical instrumentation, computerized processing of ECG has enabled consistent and fast interpretation of long-duration medical records, data archiving and development of cardiac monitoring systems. One of the prominent areas of contemporary biomedical research has been compression of ECG signals, which plays important role in applications like continuous arrhythmia monitoring, fetal ECG recording, and tele-care of elderly patients. Till date, many algorithms [1] have been proposed on ECG data compression. ECG compression algorithms employ two broad approaches: direct data compression (DDC), and transform domain (TD) methods. Among these, the DDC methods exploit the correlation between the adjacent samples (intra-beat redundancy) in a group and encode them into a smaller sub-group. Some popular algorithms employing DDC approach are AZTEC [2], turning point (TP) [3], CORTES [4], SAPA/ Fan [5, 6], interpolators [7] etc. Another method, delta encoder [8], which utilizes slope between adjacent samples in ECG, has been successfully implemented in wireless telecardiology [9]. In general, the DDC algorithms are computationally simpler and easy to implement using low end processors [10]. However, while attempting to achieve higher compression ratio, they produce serious distortion in the reconstructed ECG. Parameter extraction (PE) method, a sub-group of DDC extracts some additional features, based on the notion that consecutive ECG beats have close similarity (inter-beat redundancy). Some of the PE methods are residual-encoding [11], ECG modeling by synthesis [12, 13] etc.
TD methods, although of higher computational complexity, became popular choice for ECG compression in the last two decades. Fourier transforms [14], discrete cosine transform (DCT) [15], discrete Legendre transform [16], and Karhunen Louve transform (KLT) [17, 18] have been successfully implemented to achieve higher compression efficiency with low reconstruction error. However, discrete wavelet transform (DWT) gained maximum popularity among all TD methods for ECG compression [19–22] due to its capability to capture most of the ECG beat energy into smaller number of coefficients. The rest, insignificant coefficients are either discarded or suitably truncated using a threshold criterion for encoding (like energy packing efficiency, fixed error, data rate etc.) [23].
The performance metrics for classical (early) ECG compression works are percentage root mean squared difference normalized (PRDN), compression ratio (CR), root mean square error (RMSE) etc. defined as:
where, x, \( \overset{\frown }{x} \) and \( \overline{x} \) represent original, reconstructed and mean of original signal respectively, N: length of data array. Recently, quality of the reconstructed ECG has become an important criterion for its clinical use. It was subsequently established by researchers that metrics like PRD and PRDN can only provide a global estimate of the reconstructed signal error and have little to do with the ‘diagnostic quality’ of the ECG [24, 25]. New error measures were introduced, like weighted diagnostic distortion (WDD) [24], wavelet based diagnostic distortion measure (WWPRD) [26], etc. However, they introduce additional computational burden on the algorithm. A few wavelet based quality control approaches for ECG compression are available [26, 27]. In general, two sets of error metrics were set-forth, namely, global error measures like PRD, RMSE, SNR etc. and local distortion measure like maximum absolute error (MAE) or peak error, defined as:
This paper focused on quality criteria of ECG compressions and their control in the compression algorithm using principal component analysis (PCA). In the literature, only few works are available on KL Transform (which is similar to PCA) based ECG compression. PCA, as a powerful tool for data compression is still underutilized in ECG compression. In [18] the authors described a quality control compression based on beat segmentation, down-sampling and decomposed the ECG beats into three zones, namely, PQ, QRS and ST followed by a variance control and quality control criteria to select few basis vectors to compress the data. To the best of author’s knowledge, there has been hardly any attempt to control the compression quality by recursive selection of principal components and their adaptive adjustment of quantization level employing principal component analysis. In the proposed work, a new method is used to retain the fewer dominant principal components guided by two independent quality control criteria, namely, bit rate control (BRC) and error (or reconstruction quality) control (EC). For validation, MIT BIH Arrhythmia data (mitdb), and ECG data from PTB Diagnostic ECG data (ptbdb) under Physionet [28] were used, each of one minute duration and at 1 kHz sampling. All the results were clinically validated by two cardiologists.
Methodology
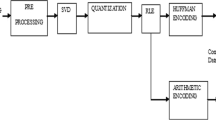
The logic flow diagram mentioning the major steps of the developed algorithm is shown by Fig. 1. The preprocessing reduces the noise and enhances inter and intra-beat correlation, which is exploited in the principal component analysis stage.
Preprocessing and beat extraction
The pre-processing stage is aimed to refine the ECG signal with the beat detection. An ECG beat is defined between two successive baseline points, detected in the TP segment between two R peaks. To achieve these, the single lead ECG data array a[] were decomposed up to 10th level using Doubechis 6 (db6) wavelet. The array was reconstructed by discarding detail coefficient D1, and D2, which represent high frequency noise and then discarding approximation coefficient A10, which represents baseline wander. QRS region is typically indicated by D4 and D5. The detail of this denoising is available in [29].
The algorithm steps for R-peak detection are given as;
/*---------------------------------------------------*/
1. x= D4×D5
2. mx_x= max(x); mn_x=min(x)
3. Threshold = 0.05× mx_x
4. Detect peaks pk[] in x[] using the Threshold
5. For each pk, find absolute maximum positions mk[] in a[] in pk±70 ms window
6. Divide the mk(k) to mk(k+1) region, bt_len(k) in 2:1
7. Detect baseline points bl[] in the region of (2/3)×bl_len±50 ms window as maximum flat region
8. Define the baseline voltage
9. Compute the amplitude of maximum (mx_x) and minimum (mn_x) points around each mk
10. Define R-peaks: if |mx_x| > 0.1× {|mx_x|+ |mn_x|}
Else, QS peak
11. Extract the beats between all bl(k) to bl(k+1) and store QRS index
/*---------------------------------------------------*/
Beat matrix generation
In this stage, beats were extracted and arranged in the form of matrix aligned with respect to the R-peak (or QS) index, with appropriate padding at the start and tails to form the beat matrix B. A single beat may be represented as:
where, b1, b2 are ECG samples, \( {\overset{\frown }{\mathrm{b}}}_k \) is the kth beat vector with j and t samples on left and right side respectively of R-peak (or QS) at the position r.
The beat matrix having m number of beats is represented as:
where, \( {\widehat{b}}_k \) is the modified beat vector having a length of l1 + l2 with (l1-j) and (l2-t) numbers of zero pads on beat-start, and beat-end respectively, l1 and l2 being the maximum beat lengths on left and right side with respect to R-peak (or QS) index.
Eigenvalue decomposition to get principal components
Principal Component Analysis (PCA) [30] is a statistical signal processing tool used to extract latent or hidden information from a multivariate data, having non-Gaussian distribution. It uses an orthogonal transformation to map the original correlated data to a set of mutually uncorrelated vectors, named principal components. These new variables are obtained in decreasing order of their variances, and hence in many cases, only few of them (i.e., less dimensions) are sufficient to represent the variability (or total energy content) of the data. These non-contributing components can be suitably truncated or eliminated without significant information loss from the original dataset. In this way, PCA can be used as a useful tool for dimensionality reduction in a correlated multivariable dataset, leading to data compression.
In the current study, the multivariate population B is R-peak-aligned beat vectors, which are mutually correlated (inter-beat correlation). A new matrix C is formed by segmenting the beat matrix B in equal partitions and using 11 beats at a time. The matrix C is then undergoes PCA after mean adjustment from each beat vector \( {\widehat{b}}_k \). The linear orthogonal transformation is represented as:
where, P = [p 1 p 2.. p m ] is the Principal Component matrix, and Ψ = [ψ 1 ψ 2 … ψ m ] is the Eigenvalues matrix.
Figure 2 represents one representative ECG beat and its energy distribution among first three PCs using PCA decomposition. It shows that the maximum energy is retained in the first PC and contributions from rest PCs are insignificant. Hence, in principle, the original data can be reconstructed by an inverse transform, retaining less number of PCs, leading to data compression.
Quality awareness in ECG compression
Quality awareness was introduced in the compression algorithms from two independent perspectives, BRC and EC. Bit Rate, expressed in bits per second, represents the post-compression number of bits used to transfer the block of data, defined as:
From Eqs. (1) to (6), we can relate the CR and bit rate. Lower the number of bits to represent compressed data, higher the CR and lower the bit rate. Hence, higher CR indicates lower bit rate. The error (reconstruction quality) was checked by PRDN and MAE in the reconstructed data, defined in Eqs. (1) and (2) respectively. From Eq. (5), the compression can be initiated by quantization of the PCs (P) and Eigenvectors matrix (Ψ), using the linear quantization formula:
Where, b = quantization level in bits, Q = quantized amplitudes of s, s min and s max are minimum and maximum values of s array respectively. The original beat B can be reconstructed by the formula:
where, Em and PCm are the modified Eigenvalues matrix and PC matrix respectively.
The final packet structure will contain compressed PCs, Eigenvectors and side information.
Error control (EC)
For EC, PRD and maximum absolute error were combined to form single evaluation criteria and used as a limit. Denoting bp: bits to represent PCs; be: bits to represent eigenvectors; N_pc: number of PCs to be retained.
The algorithm steps of EC for a single group of 11 beats are given below:
/*--------------------------------------*/
/* Error Limit set as PRDN≤ 5 % and MAE≤0.1 mV
1. N_pc=1; bp =8; be = 6;
2. Reconstruct data using Eq. (8)
3. Calculate the PRDN and MAE, discarding padding at beat-start and beat-end
4. If Error criteria is satisfied, go to step 7
5. N_pc=N_pc+1;
6. Go to step 2;
7. Compress PCs using modified delta + run length encoder or Huffman coder
8. Compress Eigenvectors using delta encoder
9. Form data packets for the current 11 beats
/*------------------------------------------*/
Bit rate control (BRC)
For BRC, the CR of a packet was put as single evaluation criteria and used as a threshold. Using the same symbols, algorithm steps are given as:
/*-------------------------------------------*/
/* Bit rate Threshold set as CR ≥ 40
1. N_pc=1; bp =8; be = 6;
2. Reconstruct data using Eq. (8)
3. Calculate the block CR, discarding padding at beat-start and beat-end
4. If CR criterion is satisfied, go to step 7
5. bp=bp -1;
6. Go to step 2
7. Compress PCs using modified Huffman coder
8. Compress Eigenvectors using delta encoder
9. Form data packets for the current 11 beats
Compression of the PCs and eigenvectors
Two different approaches were used for PC compression, based on their suitability for quality control criteria. For EC, 2–7 numbers of PCs were retained in most cases. Since variance of these lower order PCs are of low values, delta encoder followed by sign and magnitude encoding and run length encoding (RLE) was applied.
A single PC vector is represented as:
where, l is true length of the array, discarding zero pads. A down-sampling was performed to keep optimum number of samples to get array p d . The first difference was calculated as,
The array length of Δp d was adjusted by zero padding, if required, so that it becomes multiple of 8. Then, any consecutive 8-elements (say, kth group) of Δp d can be represented as,
where, skj represents sign of jth element, which is ±1, encoded as 0 or 1 respectively and |mkj| the amplitude. The encoding rule for mkj is designed as:
Finally, the RLE was applied to compress the insignificant Δp d elements is equipotential regions and lower order PCs with the following rule. The detail of the delta encoder method is given in [31].
For BRC using a single PC, Huffman coder was used on modified second difference of PC array, derives as:
The modified second difference Δp2 m was also computed. At first the unique symbols of Δp2 m were detected, followed by the construction of Huffman table and assigning of codes per symbol to construct the Huffman tree. The detail of this compression logic can be obtained in [32].
The Eigenvectors have arbitrary magnitudes. For EC, the retained eigenvectors were compressed using combination or offset logic and for BRC, the single eigenvector was retained with fixed quantization level of 6 bits.
Additional information like quantization level, number of PCs etc. was combined to form header bytes, the structure of which is given in Tables 1 and 2.
Results and discussions
The compression (and decompression) algorithm was validated using ECG data from Physionet. This website contains a large repository of various physiological signals that can be used for biomedical research. The ptbdb database under Physionet contains 12 lead ECG records with various cardiac disorders. mitdb database, which contain different types of arrhythmia records, has been universally established as benchmark for validation of compression algorithms. Since the primary objective of the proposed algorithm was to control the post-compression diagnostic quality of ECG data, four different datasets under Physionet were used: 30 sets of normal (healthy control), 15 sets each from Anterior Myocardial Infarction (AMI) and Inferior Myocardial Infarction (IMI) from ptbdb and 32 sets of Arrhythmia data from mitdb. The reason for using AMI and IMI is that they exhibit widely apart pathological symptoms [33]. For AMI records lead v4 and for IMI lead III was analyzed, where the pathologies are most prominent. For evaluation, objective and subjective measures were used. Objective test were performed by computing PRDN, CR, MAE with one minute data using Eq. (1). Subjective tests were performed by two cardiologists independently, using a double blind test.
Table 3 shows the compression performance for few arbitrarily selected AMI and IMI data. For EC with a limit of 5 % of PRDN and 0.1 mV of MAE, the average CR, PRDN, and MAE of 26.23, 4.11 and 0.057 mV respectively were found with AMI. The same figures for IMI data were 11.92, 4.88 and 0.0328 mV respectively. For BRC with a CR threshold of 40, these performance metrics came out to be 45.22, 7.90 and 0.351 mV respectively for AMI data and 48.02, 13.90, and 0.272 mV respectively for IMI data.
The reconstruction quality is represented in Fig. 3, using one each from AMI and IMI records. As expected, for EC, the reconstructed signal more closely matches with the original signal than the corresponding case in CR control, although both were clinically acceptable.
From Table 3 results it can be concluded that for BRC, the algorithm had almost similar efficiency on AMI and IMI data. For EC, CR values for IMI were low compared to AMI. The low CR value in EC is attributed to inclusion of more number of PCs and eigenvectors to meet the quality criteria. This is represented in Fig. 4, using IMI data P066/s0280lre. The entire data set was divided into 6 blocks, each consisting of 11 beats, as marked along X-axis, each ranging between a sharp peak and next lowermost valley point. The sharp peaks indicate the PRDN and MAE values with one PC and eigenvector incorporated in the reconstruction. With gradual selection of more PCs and eigenvectors, both PRDN and MAE settle down at the corresponding lowest valley point.
Table 4 shows some of the results using lead II mitdb data. For EC (PRDN limit of 5 % and MAE limit of 0.1 mV) the average CR, PRDN, and MAE were found as 9.48, 4.13 and 0.049 mV respectively, while for BRC (CR threshold at 40), these performance metrics came out to be 50.74, 16.22 and 0.243 mV respectively. In Fig. 5, the reconstruction quality for BRC and EC is shown for lead II mitdb data 117. It shows that except a few minor local distortions, the reconstructed signal closely follows the original signal plot.
The variation in PRDN and MAE with CR was investigated by varying the CR threshold in the range 40–70 using the same data and shown in Fig. 6a. It is seen that for nearly 68 % variation in CR (44 to 74), the PRDN and MAE vary only by 20 and 8 % respectively from their respective initial values. The reconstructed waveform maintains the ECG morphology (PRDN of 13.39 with MAE of 0.275 mV) till CR of 68 (Fig. 6b top panel), although some minor local distortion was observed. However, beyond CR = 70, the reconstructed morphology completely distorted (Fig. 6b bottom panel) and was clinically unacceptable.
For objective evaluation, a double blind test was performed with two cardiologists independently. They were shown the ECG plots each containing 3–4 beats, but without marking which is original and reconstructed and were requested to comment on the preservation of ECG prime clinical signatures and their diagnostic information. For EC and BRC, all the ECG data used in this study qualified for clinical acceptance.
Table 5 shows a comparison of the efficiency of the proposed work, with recent published work in similar area. It clearly indicates that the proposed method performs better than some recent published works.
Conclusion
In this paper, an algorithm for quality control in single lead ECG compression is presented using PCA with two independent approaches, namely, EC and BRC. Major contribution of the work is to retain distortion levels in the ECG within clinically acceptable limits even with high CR values (near 50). This can be useful for health monitoring applications with low link speed. For EC, the reconstruction error was very negligible. The proposed algorithm was validated with 90 data sets (60 normal and 30 diagnostic) from ptbdb and 32 datasets from mitdb. For a tighter control over reconstruction error (PRDN 5 % and MAE 0.1 mV as limit), a moderate CR of 9.48 was achieved with PRDN of 4.13 and MAE of 0.049 mV. For BRC with a target CR of 40, average PRDN, CR and MAE of 16.22, 50.74 and 0.243 mV respectively were obtained. To the best of author’s knowledge, this is the maximum average CR reported in published literature with mitdb data with acceptable reconstruction quality. For mitdb data 117, a maximum CR of 68.96 still retained ECG morphology with clinical features.
References
Jalaleddine, S. M. S., Hutchens, C. G., and Strattan, R. D., ECG data compression techniques-a unified approach. IEEE Trans. Biomed. Eng. 37(4):329–343, 1990.
Cox, J. R., Nolle, F. M., Fozzard, H. A., Oliver, G. C., and AZTEC, A preprocessing program for real-time ECG rhythm analysis. IEEE Trans. Biomed. Eng. BME-15:128–129, 1968.
Muller, W. C., Arrhythmia detection program for an ambulatory ECG monitor. Biomed Sci Instrum 14:81–85, 1978.
Abenstein, J. P., and Tompkins, W. J., New data-reduction algorithm for real-time ECG analysis. IEEE Trans. Biomed. Eng. BME-29:43–48, 1982.
Pollard, A. E., and Barr, R. C., Adaptive sampling of intracellular and extracellular cardiac potentials with the fan method. Med. Biol. Eng. Comput. 25(3):261–268, 1987.
Barr, R. C., Blanchard, S. M., and Dipersio, D. A., SAPA-2 is fan. IEEE Trans. Biomed. Eng. BME-32(5):337, 1985.
Cortman, C. M., Data compression by redundancy reduction. Proc. IEEE 133–139, 1965.
Steward, D., Dower, G. E., and Suranyi, O., An ECG compression code. J. Electrocardiol. 6(2):175–176, 1973.
Gupta, R., and Mitra, M., Wireless electrocardiogram transmission in ISM band: an approach towards telecardiology. J. Med. Syst. 38(10):1–14, 2014.
Roy, S., and Gupta, R., Short range centralized cardiac health monitoring system based on zigbee communication. Proc IEEE Global Humanitarian Technology Conference (GHTC)-South Asia Satellite (SAS), 26–27 September, 2014, Kerala, India, pp. 177–182.
Hamilton, P. S., and Tompkins, W. J., Compression of the ambulatory ECG by average beat subtraction and residual differencing. IEEE Trans. Biomed. Eng. 38(3):253–259, 1991.
Mammen, C. P., and Ramamurthi, B., Vector quantization for compression of multi-channel ECG. IEEE Trans. Biomed. Eng. 37(9):821–825, 1990.
Dutt, D. N., Krishnan, S. M., and Srinivasan, N., A dynamic nonlinear time domain model for reconstruction and compression of cardiovascular signals with application to telemedicine. Comput. Biol. Med. 33:45–63, 2003.
Al-Nashash, H. A. M., A dynamic Fourier series for the compression of ECG using FTT and adaptive coefficient. Med. Eng. Phys. 17(3):197–203, 1995.
Batista, L. V., Melcher, E. U. K., and Carvalho, L. C., Compression of ECG Signals by optimized quantization of discrete cosine transform coefficients. Med. Eng. Phys. 23(2):127–134, 2001.
Colomer, A. A., Adaptive ECG data compression using discrete legendre transform. Digital Signal Process. 7(4):222–228, 1997.
Degani, R., Bortolan, G., and Murolo, S., Karhunen Louve coding of ECG signals. Proc Computers in Cardiology, September 23–26, 1990, Chicago, pp. 395–398.
Blanchett, T., Kember, G. C., and Fenton, G. A., KLT-based quality controlled compression of single-lead ECG. IEEE Trans. Biomed. Eng. 45(7):942–945, 1998.
Xingyuan, W., and Juan, M., Wavelet based hybrid ECG compression technique. Analog Integr. Circ. Sig. Proccess. 59(3):301–308, 2009.
Chen, J., Yang, M., Zhang, Y., and Shi, X., ECG compression by optimized quantization of wavelet coefficients. Intell. Comput. Signal Process. Pattern Recogn. LNCIS 345:809–814, 2006.
Istepanian, R. S. H., Hadjileontiadis, L. J., and Panas, S. M., ECG data compression using wavelets and higher order statistics methods. IEEE Trans. Inf. Technol. Biomed. 5(2):108–115, 2001.
Kim, B. S., Yoo, S. K., and Lee, M. H., Wavelet-based low delay ECG compression algorithm for continuous ECG transmission. IEEE Trans. Biomed. Eng. 10(1):77–83, 2006.
Manikandan, M. S., and Dandapat, S., Wavelet-based electrocardiogram signal compression methods and their performances: a prospective review. Biomed. Signal Process. Control 14:73–107, 2014.
Jigel, Y., Cohen, A., and Katz, A., The weighted diagnostic distortion measure for ECG signal compression. IEEE Trans. Biomed. Eng. 47(11):1422–1430, 2000.
Al-Fahoum, A. S., Quality assessment of ECG compression techniques using a wavelet-based diagnostic measure. IEEE Trans. Inf. Technol. Biomed. 10(1):182–191, 2006.
Ku, C. T., Hung, K. C., Wu, T. C., and Wang, H. S., Wavelet based ECG data compression system with linear quality control scheme. IEEE Trans. Biomed. Eng. 57(6):1399–1409, 2010.
Alesanco, A., and Garcia, J., Automatic real-time ECG coding methodology guaranteeing signal interpretation quality. IEEE Trans. Biomed. Eng. 55(11):2519–2527, 2008.
Physionet data: http://www.physionet.org.
Banerjee, S., Gupta, R., and Mitra, M., Delineation of ECG characteristic features using multiresolution wavelet analysis method. Measurement 45(3):474–487, 2012.
Jollife, I. T., Principal component analysis. Springer, New York, 2002.
Gupta, R., and Mitra, M., An ECG compression technique for telecardiology application. Proc IEEE India Conf (INDICON), December 16–18, 2011, Hyderabad, India, pp. 1–4.
Gupta, R., Lossless compression technique for real time photoplethysmographic measurements. IEEE Trans. Instrum. Meas. 64(4):975–983, 2015.
Morris, F., Brady, W. J., and Camm, J. (Eds.), ABC of clinical cardiography, 2nd edition. Blackwell, USA, 2008.
Lee, S., Kim, J., and Lee, M., A real-time ECG data compression and transmission algorithm for an e-health device. IEEE Trans. Biomed. Eng. 58(9):2448–2455, 2011.
Mamaghanian, H., Khaled, N., Atienza, D., and Vandergheynst, P., Compressed sensing for real-time energy-efficient ECG compression on wireless body sensor nodes. IEEE Trans. Biomed. Eng. 58(9):2456–2466, 2011.
Benzid, R., Marir, F., Boussaad, A., Benyoucef, M., and Arar, D., Fixed percentage of wavelet coefficients to be zeroed for ECG compression. Electron. Lett. 39(11):830–831, 2003.
Kim, H., Yazicioglu, R. F., Merken, P., Hoof, C. V., and Yoo, H. J., ECG signal compression and classification algorithm with quad level vector for ECG holter system. IEEE Trans. Biomed. Eng. 14(1):93–100, 2010.
Mitra, M., Bera, J. N., and Gupta, R., Electrocardiogram compression technique for global system of mobile-based offline telecardiology application for rural clinics in India. IET Sci. Meas. Tech. 6(6):412–419, 2012.
Ma, J. L., Zhang, T. T., and Dong, M. C., A novel ECG data compression method using adaptive fourier decomposition with security guarantee in e-health applications. IEE J. Biomed. Health Inf. 19(3):986–994, 2015.
Acknowledgments
The author extends sincere thanks to Dr. Jayanta Saha, MD (Medicine), DM (Cardiology), and Dr. Supratip Kundu, MD (Medicine), of Calcutta Medical College & Hospital, Kolkata, India for carrying out the clinical evaluation of reconstructed data through double blind tests. The author also acknowledges SAP DRS II program 2015–2020 at Dept of Applied Physics, University of Calcutta and Department of Science & Technology, Govt. of West Bengal, India.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Systems-Level Quality Improvement
Rights and permissions
About this article
Cite this article
Gupta, R. Quality Aware Compression of Electrocardiogram Using Principal Component Analysis. J Med Syst 40, 112 (2016). https://doi.org/10.1007/s10916-016-0468-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-016-0468-7