Abstract
A pair of CuII coordination isomers [Cu(pmt)2] · 4H2O (1) and {[Cu(pmt)2] · 2H2O} n (2) [Hpmt = 2-(2-pyridylmethylamino) ethanesulfonic acid] have been prepared by reaction of the same proportion CuCl2 · 2H2O and Hpmt but with different experimental methods in water–methanol mixed solution, and were characterized by X-ray diffraction, elemental analysis, IR spectrum. X-ray analysis indicates that complex (1) is a mononuclear complex in which two deprotonated pmt ligands coordinate in a facial tridentate arrangement about the CuII atom. While complex (2) is a coordination polymer, in which four N atoms of pmt ligands coordinate to one CuII atom but its sulfonate O atoms bond to adjacent CuII atom. Thus 12-membered rings (–Cu–N–C–C–S–O–)2 are obtained between two neighbouring CuII atoms and they connect one another to form this new polymer. Crystal data: [Cu(pmt)2] · 4H2O (1), Mr = 566.10, monoclinic, P2 1/c, a = 9.1950(8) Å, b = 11.5093(10) Å, c = 11.2304(10) Å, β = 105.5550(10)°, Z = 2, V = 1144.96(17) Å3, R 1 = 0.0407, wR 2 = 0.1242 [I > 2σ (I)]; {[Cu(pmt)2] · 2H2O} n (2), Mr = 530.07, triclinic, P-1, a = 7.6165(19) Å, b = 8.806(2) Å, c = 9.592(4) Å, α = 104.933(4)°, β = 106.732(4)°, γ = 109.503(3)°, Z = 1, V = 534.2(3) Å3, R 1 = 0.0470, wR 2 = 0.1082 [I > 2σ(I)].
Index Abstract
A pair of CuII coordination isomers [Cu(pmt)2] · 4H2O (1) and {[Cu(pmt)2] · 2H2O} n (2) [Hpmt = 2-(2-pyridylmethylamino) ethanesulfonic acid] have been prepared by reaction of the same proportion CuCl2 · 2H2O and Hpmt but with different experimental methods in water–methanol mixed solution.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Schiff base complexes containing both sulfur and amino acid functionalities have aroused considerable recent interest because of their biological activities [1–3]. 2-Aminoethanesulfonic acid, known as taurine, is indispensable to human life and plays a significant part in physiological functions. Casella and Gullotti [4] have found that zinc and copper complexes of schiff bases ligands, prepared by the condensation of 2-pyridinecarboxaldehyde with non-polar amino acid, are unstable. This has been attributed to the rigidity of the C=N double bond in the free schiff bases and this agreement is later confirmed by Wagner and Walker [5]. In order to explore complexes that are more stable and have much stronger biological activities, we have recently drawn our attention to coordination complexes based on the reduced schiff base ligand 2-(2-pyridylmethylamino) ethanesulfonic acid (Hpmt). The present paper reports the syntheses and crystal structures of two CuII coordination isomers [Cu(pmt)2] · 4H2O (1) and {[Cu(pmt)2] · 2H2O} n (2), whose X-ray analysis reveals that (1) is a mononuclear structure and (2) is a coordination polymer.
Experimental Section
All chemicals except the Hpmt ligand were of reagent grade obtained from commercial sources and used without further purification. The C, H, N, S elemental analyses were preformed on a Elementar Vario EL elemental analyzer. The IR spectrum was recorded with a Shimadzu IR-408 spectrophotometer using the KBr pellet in the range of 4,000–400 cm−1. The Hpmt ligand was prepared according to the procedure in literature [6].
Synthesis of [Cu(pmt)2] · 4H2O (1)
A solution (5 ml) of ligand Hpmt (2 mmol, 0.432 g) was basified with KOH (1 mol/l) to a pH of 7.5–8.0. Then a solution of CuCl2 · 2H2O (0.171 g, 1.0 mmol) in methanol (8 ml) was added dropwise to it. The resulting mixture was stirred at 333 K for 6 h, filtrated, and kept at room temperature to obtain blue, block-shaped crystals after 1 week. Yield: 0.26 g (47%). Anal. Calcd for C16H30CuN4O10S2: C, 33.92; H, 5.30; N, 9.89; S, 11.30%. Found: C, 33.87; H, 5.33; N, 9.92; S, 11.25%. IR data (cm−1): 3482 (s), 3232 (m), 1612 (m), 1573 (m), 1217 (s), 1165 (s), 1041 (s), 771 (s), 747 (m).
Synthesis of {[Cu(pmt)2] · 2H2O} n (2)
A solution (5 ml) of ligand Hpmt (2 mmol, 0.432 g) was added dropwise to a solution of CuCl2 · 2H2O (0.171 g, 1.0 mmol) in methanol (8 ml), the obtained mixture was stirred at 333 K for 3 h. After that, the mixture was basified with KOH (1 mol/l) to a pH of 7.5–8.0 and continued stirring for another 6 h, filtrated. One month later, dark-blue block crystals were grown from the filtrate by slow evaporation. Yield: 0.18 g (34%). Anal. Calcd for C16H26CuN4O8S2: C, 36.26; H, 4.91; N, 10.57; S, 12.08%. Found: C, 36.22; H, 4.96; N, 10.61; S, 12.01%. IR data (cm−1): 3426 (s), 3233 (m), 1614 (m), 1570 (m), 1213 (s), 1185 (s), 1037 (s), 781 (s), 726 (m).
X-ray Diffraction Determination
The diffraction data for (1) and (2) were collected on a Bruker APEX-II CCDC areadetector diffractometer using graphite-monochromatized Mo-K α radiation (λ = 0.71073 Å). Cell constants and orientation matrix for data collection were obtained from least-squares refinement by using the setting angles in the range of 2.58–27.50 for (1) and 2.41–25.50 for (2). Absorption corrections were applied using the SADABS program [7]. The structure was solved by direct methods [8] and refined by full-matrix least squares on F 2 using the SHELXL97 software [9]. All the non-hydrogen atoms were anisotropically. The hydrogen atoms were generated geometrically and treated by a mixture of independent and constrained refinement. The crystallographic data for complexes 1–2 are listed in Table 1.
Results and Discussion
The thermal ellipsoid plots for (1) and (2) with the percent probability of 30% are depicted in Figs. 1 and 2, respectively. The selected bond lengths (Å) and angles (°) are shown in Table 2.
Description for [Cu(pmt)2] · 4H2O (1)
The crystal structure reveals that compound (1) is a mononuclear complex which is comprised of one CuII atom, two deprotonated pmt anion ligands and four water molecules. As displayed in Fig. 1, the CuII atom lies on an inversion center with the two pmt ligands coordinated in a tridentate trans arrangement with pyridyl N, amino N and sulfonate O atoms as donors (Fig. 1). The coordination environment at the Cu is distorted octahedral. This complex is isostructural with [Co(C8H11N2O3S)2] whose structure has been determined earlier [6]. The bond length of Cu–O [2.398(2) Å] is much longer than that of Cu–N [(2.039(2) Å, 2.042(2) Å], while in Co complex they are almost equal [Co–O, 2.1004(11) Å; Co–N, 2.1604(12) Å, 2.1737(13) Å]. This difference shows that there is Jahn-Teller effect for Cu and lack of Jahn-Teller for Co.
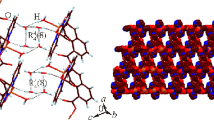
The water molecules in the lattice and the S=O groups of the pmt ligand are involved in the hydrogen bonding and form three-dimensional network (Fig. 3) These intermolecular hydrogen bonds are of medium strength as the D···A distances are in the range of 2.773(7)–2.923(4) Å, tabulated in Table 3.
Moreover, there exists weak π–π stacking interactions between the parallel pyridine rings of the adjacent units (Fig. 4). The interplanar perpendicular distance and ring-centroid separation distance are 3.528(5) and 4.046(1) Å, respectively. π–π stacking help to stabilize three-dimensional network.
Description for {[Cu(pmt)2] · 2H2O} n
The crystal structure indicates that compound (2) is a coordination polymer which is build up from one CuII atom, two pmt ligands and two water molecules. As shown in Fig. 2, the CuII atom is situated on a center of symmetry and is six-coordinated with four N atoms from two pmt ligands together with two sulfonate O atoms belonging to another two ligands, exhibiting a distorted octahedral geometry. Four N atoms occupy the equatorial plane and they are coplanar and O atoms locate on axial position. The distance of Cu–O bond [2.572(2) Å] is evidently longer than that of Cu–N bond [2.011(3) and 2.039(3) Å], showing the typical Jahn-Teller distortion effect for the CuII atom.
In complex 2, the N atoms of each pmt ligand bond to one CuII atom, but its O atom coordinates to adjacent CuII atom. This original μ 2-bridging coordination mode leads to forming 12-membered (–Cu–N–C–C–S–O–)2 rings between neighbouring CuII atoms. These interconnected rings create a one-dimensional linear chain propagating along the a axis in the triclinic unit cell, with a Cu···Cu distance of 7.617(2) Å.
Stronger π–π stacking interactions can be observed between the parallel pyridine rings of adjacent chains along a axis (Fig. 5). The interplanar average distance and ring-centroid separation distance are 3.455(4) and 3.710(6) Å, respectively. The chain structure is expanded into two-dimensional network in the ab plane by π–π stacking.
In addition, there exists weak hydrogen bonding interactions between lattice water molecules and pmt ligand. They connect together to form eight-membered (H–N–Cu–O–S–O–H–O) and 12-membered (–O–S–O–H–O–H–)2 rings between adjacent chains (Fig. 6), which join the network into a three-dimensional supramolecular architecture.
Conclusions
In this paper, we report the syntheses and structures of a pair of new copper coordination isomers [Cu(pmt)2] · 4H2O (1), {[Cu(pmt)2] · 2H2O} n (2). It is just the using of different experimental methods that result in the pmt ligand coordinates to CuII atom in different modes: pmt in (1) can be described as N,N′,O-tridentate chelating coordination, signed as “3” mode; pmt in (2) is N,N′-copper, O-another copper, marked as “2 + 1”mode. Both the hydrogen bonds and π–π interactions make great contributions on stabilization the three-dimensional structure.
Supplementary Data
CCDC numbers 614217 and 629383 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html [or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge, CB2 1EZ, UK; Telephone: (44) 01223 762910; Facsimile: (44) 01223 336033; E-mail: deposit@ccdc.cam.ac.uk].
References
Casella L, Gullotti M (1981) J Am Chem Soc 103:6338
Casella L, Gullotti M (1986) Inorg Chem 25:1293
Wang Z, Wu Z, Yen Z, Le Z, Zhu X, Huang Q (1994) Synth React Inorg Met Org Chem 24:1453
Casella L, Gullotti M (1983) Inorg Chem 22:2259
Wagner MR, Walker FA (1983) Inorg Chem 22:3021
Li JX, Jiang YM, Li HY (2006) Acta Crystallogr E62:m2984
Sheldrick GM (1996) SADABS, program for absorption correction of the area detector data. University of Göttingen, Germany
Sheldrick GM (1997) SHELXS97, program for crystal structure solution. University of Göttingen, Germany
Sheldrick GM (1997) SHELXL97, program for crystal structure refinement. University of Göttingen, Germany
Acknowledgements
This work was funded by the Guangxi Science Foundation of the Guangxi Zhuang Autonomous Region of the People’s Republic of China (Grant No. 0731053). We also thank Luoyang Normal College and Nankai University for supporting this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, JX., Jiang, YM. & Lian, BR. Syntheses and Crystal Structures of Two CuII Coordination Isomers [Cu(pmt)2] · 4H2O (1) and {[Cu(pmt)2] · 2H2O} n (2). J Chem Crystallogr 38, 711–715 (2008). https://doi.org/10.1007/s10870-008-9365-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10870-008-9365-3