Abstract
Raman spectroscopy has been used to determine the chemical composition of materials for over 70 years. Recent spectacular advances in laser and CCD camera technology creating instruments with higher sensitivity and lower cost have initiated a strong resurgence in the technique, ranging from fundamental research to process control methodology. One such area of increased potential is in tissue engineering and regenerative medicine (TERM), where autologous cell culture, stem cell biology and growth of human cells on biomaterial scaffolds are of high importance. Traditional techniques for the in vitro analysis of biochemical cell processes involves cell techniques such as fixation, lysis or the use of radioactive or chemical labels which are time consuming and can involve the perpetuation of artefacts. Several studies have already shown the potential of Raman spectroscopy to provide useful information on key biochemical markers within cells, however, many of these studies have utilised micro- or confocal Raman to do this, which are not suited to the rapid and non-invasive monitoring of cells. For this study a versatile fit-for-purpose Raman spectrometer was used, employing a macro-sampling optical platform (laser spot size 100 μm at focus on the sample) to discriminate between different TERM relevant cell types and viable and non-viable cells. The results clearly show that the technique is capable of obtaining Raman spectra from live cells in a non-destructive, rapid and non-invasive manner, however, in these experiments it was not possible to discriminate between different cell lines. Despite this, notable differences were observed in the spectra obtained from viable and non-viable cells, showing significant changes in the spectral profiles of protein, DNA/RNA and lipid cell constituents after cell death. It is evident that the method employed here shows significant potential for further utilisation in TERM, providing data directly from live cells that fits within a quality assurance framework and provides the opportunity to analyse cells in a non-destructive manner.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Tissue Engineering and Regenerative Medicine (TERM) is a rapidly emerging interdisciplinary field of research involving the life, physical and engineering sciences that seeks to replace or regenerate human tissue or organs, to restore or re-establish normal bodily function [1]. For regenerative medicine to be successful it requires a capability to expand relevant cell populations such as Mesenchymal stem cells (MSC), Overcoming the economic, technical, regulatory and quality assurance challenges that exist as barriers to progress in the area is currently seen as critical for benefits to reach patients. Of particular interest here is the ability to monitor cell status (viability and phenotype) during culture in a non-destructive, rapid and non-invasive manner. Currently there are a number of methods available to monitor the in vitro interactions that occur between cells with their environment, including those based on microscopy, immunochemical and molecular biochemical techniques. However, whereas there is merit in each, none effectively provide for these core criteria [2]. The techniques that are routinely used by cell biologists, are generally invasive, and require fixation of the cells, the use of molecular probes or lysis of the cells [2, 3]. In the case where cell viability and actual phenotype are under investigation these methods used are obviously destructive and therefore do not suit circumstances where the expanded cell cultures are required to be used directly for clinical implantation. Hence it is necessary to develop alternative strategies for in situ and non-destructive analysis. To this end, a number of alternative techniques are currently being investigated to analyze cells and tissues for tissue engineering, including, surface plasmon resonance (SPR), bioluminescence imaging (BI), optical coherence tomography (OCT), Fourier Transform Infrared Spectroscopy (FTIR) and Raman Spectroscopy [4]. Of these techniques, Raman spectroscopy has shown particular promise [2, 3] as evidenced by its increasing utility in the life sciences sector in recent years [5–17]. One of the drivers for this transformation has been the evolution of the instrumentation to the stage where the technique’s potential can be realised in complex solutions. Seminal research by Puppels et al. demonstrated the efficacy of using Raman Spectroscopy to investigate live cells without the need for complex in vitro assays [11]. More recently, microscope based systems employing a near infrared (NIR) laser source have been successfully utilized in a range of studies involving living cells [5–8]. These include investigations into cell death [5, 14], phenotypic identification of cells [7], proliferation [10], mitosis [13], and differentiation [9]. Although extremely useful, none of these approaches really lend themselves to the requirements for in-situ, real-time, rapid and non-invasive monitoring of cells in their current configurations. As such, this study aims to utilise a versatile fit-for-purpose Raman optical platform capable of delivering data directly from live cells that fits within a quality assurance framework and provides the opportunity to analyse cells in a non-destructive manner. The purpose of this study was to examine a series of well characterised cell lines using a Bench-top Macro-Raman spectrometer with high throughput screening (HTS) capability. The system in question would utilise an optical platform with a 100 μm laser spot size at focus on the sample to investigate the differences between different cell lines and the effects of cellular death.
2 Materials and methods
2.1 Cell culture
For this work, a range of TERM relevant cell lines were cultured under their specific culture conditions prior to investigation using Raman spectroscopy and Immunocytochemistry. All cells were cultured under standard conditions of 37°C, and 95% air and 5%CO2 in T75 culture flasks (Nunc, UK). All reagents used were from Invitrogen, Paisley, Scotland unless otherwise stated). Immortalised cell lines were chosen in this case as they have already been well characterised and represent a suitable benchmark before proceeding to primary and stem cell lines.
2.1.1 SAOS2 osteosarcoma cell line
The human osteoblast cell line SAOS2 derived from bone osteosarcoma cells were purchased from ATCC Global Bioresource Center. The cells were cultured in McCoy’s 5a medium (modified) with 1.5 mM L-glutamine adjusted to contain 2.2 g/l sodium bicarbonate. Media was supplemented with 15% fetal bovine serum containing 100 units/ml of penicillin, 100 μg/ml of streptomycin sulphate and 25 μg/ml of amphotericin B.
2.1.2 WS1 human dermal fibroblast cell line
The normal human dermal fibroblast cell line WS1 (CRL-1502) was purchased from the ATCC (Global Bioresource Center). Cells were cultured in Minimum Essential Medium (Eagle’s) supplemented with 10% fetal calf serum containing 100 units/ml of penicillin, 100 μg/ml of streptomycin and 25 μg/ml of amphotericin B. All reagents used were from Sigma Chemicals Ltd.
2.1.3 Human embryonic lung epithelial L132 cells
Human embryonic lung epithelial L132 cells were purchased from the ATCC Global Bioresource Center were propagated in Minimum Essential Medium (Eagle) with 2 mM L-glutamine and Earle’s BSS adjusted to contain 1.5 g/l sodium bicarbonate, 0.1 mM non-essential amino acids, and 1.0 mM sodium pyruvate, supplemented with 10% fetal bovine serum containing 100 units/ml of penicillin, 100 μg/ml of streptomycin sulphate and 25 μg/ml of amphotericin B.
2.1.4 MG63 cell line
The MG-63 cells derived from an osteosarcoma of a 14-year-old male were purchased from the ATCC (Global Bioresource Center). Cells were cultured in Minimum Essential Medium (MEM) supplemented with 10% fetal calf serum and antibiotic/antimycotic (penicillin G sodium 100 U/ml, streptomycin 100 μg/ml, amphotericin B 0.25 μg/ml, (PAA Laboratories GmbH, Austria).
2.1.5 Immunocytochemistry
In order to ascertain whether the different cell lines were living or dead, samples of each were cultured on glass coverslips under normal conditions until they became pre-confluent and cell damage was then induced by incubation of slides with fresh media containing 100 μM Triton X-100 for 24 h. Immunocytochemistry was performed to examine the morphology of the cells. Briefly, cell cultures were grown for 24 h following passage and cells rinsed in ice-cold PBS and fixed and permeabilised in 3.7% Paraformaldehyde containing 0.1% Triton X-100 for 20 min at 4°C. The cells were then washed in PBS to remove traces of the PFA (3 × 15 min) prior to staining of the cell actin cytoskeleton with Alexa Fluor 488 phalloidin probe (Molecular Probes, Inc. Invitrogen) for 20 min. After washing twice in PBS (2 × 5 min), the slides were mounted in glycerol mounting media containing anti-fade, sealed using nail varnish and the cells were then viewed using a Carl Zeiss LSM5 Pascal confocal scanning laser microscope with dual lasers.
2.2 Analysis of the cells
In order to analyse the cells using a Raman spectrometer, the relevant cell lines were grown to confluence as outlined in Sect. 2.1. The cells were then trypsinized (0.25% trypsin/EDTA), centrifuged at low speed and re-suspended in media for 1 hour. After this, the suspension was centrifuged at 5,000 rpm in order to produce a cell pellet suitable for analysis using the Raman spectrometer. As well as analysing the different cell lines, live cell pellets and cell pellets that were subjected to Triton X-100 for 24 h (to induce cell death) were also analysed (as detailed in Sect. 2.2.1). Cell pellets were placed on fused silica and analysed immediately using the Raman spectrometer. All cell lines were analysed in triplicate by utilising the automated sampling routines available in the Raman spectrometer software (Insight, Avalon Instruments, Belfast).
2.2.1 Induction of cell death
In order to induce cell death in cell populations prior to Raman analysis, samples of the various cell lines in this study were incubated in 100 μM Triton X-100 for 24 h. These samples were analysed using Macro-Raman spectroscopy (laser spot size of 100 μm) alongside samples of the different cell lines that were cultured under normal conditions as outlined previously in Sect. 2.1.
2.2.2 Raman spectroscopy
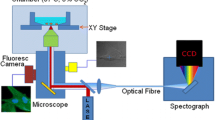
Raman spectroscopy was performed on the cell pellets using an Avalon RamanStation, Belfast, Northern Ireland) equipped with a stabilised 785 nm near-infrared (NIR) diode laser and an Echelle spectrograph allowing for the full spectrum to be analysed without any moving parts. Laser power was 100 mW at the sample. Pellets of each cell type were analysed in triplicate using an autofocus routine for each sample to ensure optimal sample positioning and signal-to-noise ratio. The instrument was configured for macro-sampling (in order to account for sample inhomogeneity), allowing the collection of Raman scattering from a large area (100 μm diameter spot). This provides the advantage of analysing a large number of cells at the same time as opposed to micro-Raman spectroscopy that analyses only a single cell. Scans were recorded from 3400 to 250 cm−1 with up to a 60 second exposure time. The instrument was wavelength calibrated using a neon source and verified using a polystyrene secondary standard (American Society for Testing and Materials (ASTM) E 1840-96). The spectra were also intensity corrected using the National Institute of Standards and Technology (NIST) Standard Reference Material® (SRM) 2241. The Data obtained were processed by incorporating a systematic multipoint baseline correction across all spectra using GRAMS AI (Thermo Galactic). All spectra were normalised using the peak at 1450 cm−1, which corresponds to C–H vibrations from all the cellular components [5, 14].
3 Results and discussion
3.1 Cell morphology & immunofluorescence
Prior to Raman analysis the different cell lines cultured in the T75 flasks were examined using phase contrast microscopy to ensure that they exhibited typical cell morphology and that the cell populations were viable. Figure 1a and c shows typical phase contrast images of exemplar L132 Lung epithelial cells and SAOS2 osteoblast like cells that were cultured under normal conditions (and in the absence of Triton-X-100). As was the case for all the cell lines investigated here, the cells exhibited the expected cell morphology at both pre-confluency and confluency. However, for those flasks that were treated with Triton-X-100, the number of adherent cells on the flask were significantly reduced as is shown in Fig. 1b and d for the L132 and SAOS2 cell, respectively. There was also significant rounding and shrinking of the cells observed here, with a significant number of cells seen to be floating in the media. This indicates that the Trinton-X-100 has induced cell death in the population as compared to those cells not exposed and cultured under normal conditions.
Further experiments were conducted to ascertain the viability of the various cell populations (with and without exposure to Trinton-X-100) using Immunocytochemistry. All cell types cultured on glass coverslips demonstrated the typical cell morphology expected in the absence of Triton-X-100 at both pre-confluency and confluency as shown in Fig. 2a and c for the L132 and SAOS2 cell lines, respectively. Immunofluorescence localisation of actin expression in permeabilised cells demonstrated a distinctive change in actin cytoskeletal architecture in cells incubated in the presence of Triton X-100 for 24 h, as shown in Fig. 2b and d for the L132 and SAOS2 cell lines, respectively. This provides a clear indication, more so than the phase contrast images, that the Trinton-X-100 has induced cell death at 24 h. For those cells exposed to the Trinton-X-100, there is clear evidence of cell smearing and membrane blebbing, with significant changes in the cell morphology and cell definition indicative of membrane disruption.
As such, the experiments conducted here provides confidence that the cells that have been subjected to Trinton-X-100 are dead, whereas those cells cultured under normal conditions (without exposure to Trintox-X-100) are viable and can go forward to be analysed using Raman Spectroscopy. It should be noted that all cell lines tested exhibited the same results as those reported here for the L132 and SAOS2 cell lines using phase contrast microscopy and Immunocytochemistry techniques.
3.2 Raman spectroscopy of the different cell lines
Raman spectra for healthy viable cells are shown in Fig. 3a–d for the L132, WS1, MG63 and SAOS2 cell lines, respectively. They all indicate significant contributions from protein, nucleic acid, lipid and carbohydrate components and are comparable to those observed previously by Notingher et al. [5, 14]. These include typical protein peaks, such as those observed at 645 cm−1 and 854 cm−1 for Tyrosine and 623 cm−1 and 1005 cm−1 for Phenylalanine. Other typical protein peaks are observed between 1128–1450 cm−1 and 1600–1660 cm−1 as expected [5, 10, 14]. Peaks indicative of RNA and DNA are clearly observed at 782, 788, 1095, 1320–1342 and 1572 cm−1, as would be expected, however, several key peaks corresponding to RNA and DNA were absent from the spectra, such as those at 897 and 1013 cm−1 [5, 14, 17]. Lipid peaks are also observed in all of the spectra for the viable cells between 1066–1128, 1260–1300, 1449 and 1660 cm−1 [5, 10, 14]. Again, several peaks indicative of lipids were not clearly observed, particularly those normally found at 1367 and 1460 cm−1 [5, 14]. The absence of such peaks could be as a result of numerous factors, including a lack of sensitivity given the optical platform employed for these experiments, differences due to when the cells were analysed in their respective cell cycles, damage to the cells on the outer surface of the pellet, and problems related to focusing the laser spot on the cell pellet (or combinations of these factors). Peaks indicative of carbohydrates can also be seen around 1080–1095 cm−1 [5, 14] along with significant C–H stretching bands (from mainly lipids and proteins) between 2800 and 3050 cm−1 [10]. The spectra obtained from the different cell lines all show distinct similarities in the peak positions of their various cellular components, however, it is important to note that there are some observable differences in their spectral profiles. Most notable are those difference seen around 800–850, 1200–1300 and 1500–1600 cm−1. As many of these particular areas are associated with protein and nucleic acid peaks it is suggested that these differences in the spectra are more likely to be a consequence of the cells being in different phases of their life cycle as opposed to observable biochemical differences between different cell lines. Similar observations were made by Notingher et al. previously [5, 14, 17]. Indeed, the experiments conducted here were not contrived to ensure that the cells were at a particular phase of the cell cycle and as such the results here can only show the spectra of live cells and cannot be utilised to discriminate between different cell lines.
3.3 Raman spectroscopy for monitoring cell viability
Raman spectra for both the viable and non-viable MG63 Osteoblast like cell line are shown in Fig. 4a and b respectively. These spectra clearly show significant differences between the viable healthy cells (no Trinton-X-100 treatment) and the non-viable cells (Trinton-X-100 treated) as would have been expected. Notable spectral differences are observed between 770 and 900 cm−1. Most of the main peaks observed here for the live cells were not detected for the Trinton-X-100 treated cells. Normally peaks in this area are associated with DNA and RNA. In particular, the peak observed at 788 cm−1, normally associated with the O–P–O stretching of DNA phosphodiester bonds and the peak at 782 cm−1 corresponding to the ring stretching of cytosine and thyamine [5, 10, 14]. In particular, the loss of intensity of the O-P-O stretching of DNA Phosphodiester bonds at 788 cm−1 indicates breaking of the DNA strands, which is also confirmed by the absence of the peak at 828 cm−1. The peak at 1572 cm−1 corresponding to DNA and RNA also decreases in intensity after exposure to Trinton-X-100 which again confirms this hypothesis. However, no significant decrease was observed in the phenylalanine peak at 1005 cm−1 or the nucleic acid peak at 1095 cm−1 as a result of cell death, which has been previously observed by others [5, 14]. In the case of the peak at 1095 cm−1, This may be due to the fact that this peak can also be assigned to lipids, proteins and carbohydrates and therefore changes in this particular peak may not be significant. A significant decrease was observed in the intensities of those bands associated with proteins between 1280 and 1350 cm−1, which confirms the observations made previously by Notingher and is a clear indication of cell death [5, 14]. There was a significant broadening of the large peak between 1570 cm−1 and 1730 cm−1, particularly on the lower wavenumber side. This could possibly indicate an increase in the presence of aromatic amino acids as a result of protein cleavage from cell death.
There was a significant increase in the intensity of peaks observed between 1020 and 1080 cm−1 as a consequence of exposure to the Trinton-X-100. Peaks in this area can be associated with lipids [5, 14]. Further evidence for this increase in lipid concentration is provided by the increase in the intensity of the broad band between 1220 and 1300 cm−1 associated with =CH deformations in lipids and the band at 1660 cm−1 (normally attributed to C=C stretching) however, other peaks associated with lipids did not show any significant changes in intensity. This is most notable for the peak at 1301 cm−1, where no significant change in intensity was observed. No lipid peak was observed at 1743 cm−1 for either the live or dead cells in these experiments as was the case for other published work [5, 14]. A further peak at 1486 cm−1 was also observed in the spectrum for the non-viable cells, however, due to the complex nature of the spectra obtained from the cells it could not be assigned to a particular cellular component at this time. In general, the results obtained from the different cell lines exposed to the Trinton-X-100 solution for 24 h were very similar to those observed for the MG63 osteoblast like cell line and were not considered significant.
The results present here clearly indicate that Raman systems that are not configured for micro- or confocal Raman spectroscopy, such as the versatile system employed here, can be utilised to monitor cell behaviour in real-time and in non-destructive and non-invasive manner. However, it is evident from the data obtained here and the observations from Notingher [5, 14], that in order to clearly discriminate between different cell lines using such techniques (in a research context) it would be preferable to ensure that the cells were at the same phase of the cell cycle. In the first instance it will be important to have benchmark spectra indicative of a particular cell line at the various phases of its cell cycle, and to understand the peak assignments associated with the individual cellular components during each phase before translating such technology into a clinical or manufacturing setting. However, in a real application area, such as the expansion of a cell population for use in a TERM product, it is unlikely that all of the cells would be at the same phase of the cell cycle all of the time and as such, any method used to analyse the cells should be capable of coping with such situations. Certainly it is considered that the versatile Raman platform utilised here, incorporating a larger laser spot size for analysis than previously published studies is more appropriate for process monitoring applications, such as those considered necessary in the regulatory framework required for TERM products. The system takes an average spectrum from a large number of cells rather than analysing individual cells (or areas within individual cells), and as such is more representative of the cell population as whole. It also provides a technique to analyse a larger number of cells more rapidly than before. Previous authors required spectral acquisition times of between 2 and 10 min per cell, whereas the approach taken here could analyse multiple cells in less than 60 s. Indeed, acquisition times of less than 30 s could provide usable spectra from live cells.
A further observation from the analysis of live versus dead cells shows that the versatile Raman platform utilised here can easily discriminate between the two cell states. However, in the experiments conducted here, some of the key spectral differences that were utilised by previous authors to discriminate between live and dead cells were not observed, Most notably, no significant difference was observed in the intensity of the phenylalanine peak at 1005 cm−1 between the live and dead cells. This is despite the decrease in intensity of other key peaks associated with proteins after cell death (1280–1350 cm−1, the increase in the intensity of some of the key lipid peaks (1020–1080, 1220–1300 and 1660 cm−1) and the loss of key DNA/RNA peaks (788 cm−1), all being clearly evident. These changes in the spectral profile for these various cellular components have all been identified as key Raman spectral markers of cell death by previous authors [5, 14]. The spectral changes observed as a result of cell death are the effects of the complex molecular mechanisms during the induction of cell death, such as protein cleavage due to the activation of caspases, followed by DNA fragmentation. As such, the absence of any significant difference in the peak associated with phenylalanine (1005 cm−1) after cell death cannot be taken in isolation, given the other significant differences observed across the entire spectral profile. Indeed, previous work by others observed similar instances whereby key markers of cell viability or non-viability were not observed. This may be explained by the non-homogeneous distribution of the various cellular components in the cell pellet or the fact that the cell population could possibly contain a significant population of viable cells, despite the exposure to Trinton-X-100 for 24 h.
4 Conclusion
This study clearly highlights the versatility of using a Raman spectrometer that incorporates a larger laser spot size for the analysis of viable cell populations. It is evident from the work presented here that this can be achieved using a Raman system that is not microscope based and is suited to high throughput screening of large populations of cells. The analysis times are significantly reduced, providing data that can clearly discriminate between viable and non-viable cell populations. As yet, the Raman platform utilised here cannot discriminate between different cell lines, however, further experimentation is planned in order to address this shortfall in the technique, including the use of statistical models. Despite this, the system employed here provided Raman data in real time, in a non destructive manner and non-invasive manner (without the need for staining). As yet it is not optimised for utilisation in a clinical or manufacturing environment for the expansion of healthy cell populations. However, it does show significant potential for utilisation in TERM applications and is the subject of further detailed investigations by the authors.
References
Mason C, Dunhill P. Regen Med. 2008;3:1–5.
Notingher I, Hench LL. Expert Rev Med Devices. 2006;3:215–34.
Swain RJ, Stevens MM. Biochem Soc Trans. 2007;35:544–9.
Mather ML, Morgan SP, Crow J. Regen Med. 2007;2:145–60.
Notingher I, Verrier S, Haque S, Polak JM, Hench LL. Biopolymers (Biospectroscopy) 2003;72:230–40.
Notingher I, Bisson I, Polak JM, Hench LL. Vib Spec. 2004;35:199–203.
Notingher I, Jell G, Lohbauer U, Salid V, Hench LL, Cell J. Biochemistry. 2004;92:1180–92.
Notingher I, Bisson I, Bishop AE, Randle WL, Polak JM, Hench LL. Anal Chem. 2004;76:3185–93.
Notingher I, Jell G, Bisson I, Polak JM, Hench LL. Bioceramics. 2005;17:545–8.
Short KW, Carpenter S, Freyer JP, Mourant JR. Biophys J. 2005;88:4274–88.
Puppels GJ, Mul FF, Otto C, Greve J, Robert-Nicoud M, Arndt-Jovin DJ, et al. Nature. 1990;347:301–3.
Jeannesson P, Manfait M, Jardillier JC. Anal Biochem. 1983;129:305–9.
Matthäus C, Boydston-White S, Miljković M, Romeo M, Diem M. Appl. Spectrosc. 2006;60:1–8.
Verrier S, Notingher I, Polak JM, Hench LL. Biopolymers. 2004;74:157–62.
Xie C, Goodman C, Dinno MA, Li Y. Opt Express. 2004;12:6208–14.
Uzunbajakava N, Lenferink A, Kraan Y, Willekens B, Vrensen G, Greve J, et al. Biopolymers (Biospectroscopy). 2003;72:1–9.
Owens CA, Notingher I, Hill R, Stevens M, Hench LL. J Mat Sci Mat Med. 2006;17:1019–23.
Acknowledgements
The authors would like to express their gratitude to the EPSRC for funding this project (EP/C534247/1—Remedi—Regenerative Medicine—A New Industry).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boyd, A.R., Burke, G.A. & Meenan, B.J. Monitoring cellular behaviour using Raman spectroscopy for tissue engineering and regenerative medicine applications. J Mater Sci: Mater Med 21, 2317–2324 (2010). https://doi.org/10.1007/s10856-009-3965-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10856-009-3965-0