Abstract
We calculate the absolute binding free energies of tetra-methylated octa-acids host–guest systems as a part of the SAMPL6 blind challenge (receipt ID vq30p). We employed two different free energy simulation methods, i.e., the umbrella sampling (US) and double decoupling method (DDM). The US method was used with the weighted histogram analysis method (WHAM) (US-WHAM scheme). In the DDM scheme, Hamiltonian replica-exchange method (HREM) was combined with the Bennett acceptance ratio (BAR) (HREM-BAR scheme). We obtained initial binding poses via molecular docking using GalaxyDock-HG program, which is developed for the SAMPL challenge. The root mean square deviation (RMSD) and the mean absolute deviations (MAD) using US-WHAM scheme were 1.33 and 1.02 kcal/mol, respectively. The MAD was the top among all submissions, however the correlation with respect to experiment was unexceptional. While the RMSD and MAD via HREM-BAR scheme were greater than US-WHAM scheme, (i.e., 2.09 and 1.76 kcal/mol), their correlations were slightly better than US-WHAM. The correlation between the two methods was high. Further discussion on the DDM method can be found in a companion paper by Han et al. (receipt ID 3z83m) in the same issue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ability to predict binding affinities of protein–ligand has been a longstanding goal of computational chemists and biologists. An accurate prediction can accelerate the challenging process of designing and optimizing a new drug candidate [1, 2]. For example, binding affinity prediction based on molecular simulations are used to virtual screening, evaluating target toxicity and potential side-effects of leads or drug candidates.
Host–guest systems are useful model for validating computational methods for predicting protein–ligand binding affinities. It significantly reduces the complexity and cost of computations. Host molecules used in SAMPL challenges are smaller (a few hundred atoms) than proteins but retaining cavities or clefts which are large enough to bind to drug-like small molecules. As host molecules are more rigid and have fewer degrees of freedom than proteins, random error due to uncertainty in sampling can be dramatically reduced. In fact, host–guest systems have attracted great attention in pharmaceutical sciences, biology, chemistry, and nanotechnology, enabling “bottom-up” approach for understanding intricate protein–ligand interactions.
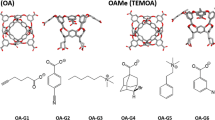
Host–guest systems have been included in the Statistical Assessment of the Modeling of Proteins and Ligands (SAMPL) blind challenge [3,4,5,6,7,8,9,10,11] since SAMPL3 in 2011. Octa-acids (OA) [12] and tetra-methylated octa-acids (TEMOA) [13], which are previously known as OAH and OAMe, respectively, have also been introduced in the SAMPL4 [13, 14] and the SAMPL5 [15] challenges. Both molecules were developed by Gibb and co-workers. The two hosts are identical except that TEMOA has for additional methyl groups, which alter the shape and depth of the hydrophobic cavity. The two hosts are completely identical except that TEMOA has for additional methyl groups, which alter the depth of the hydrophobic cavity, while OA has hydrogen atoms at the parts. For the sixth edition of the SAMPL (SAMPL6), Gibb and co-workers provided the binding free energy values, measured by ITC, for eight guests interacting with OA and TEMOA. The measurements were performed in 10 mM sodium phosphate buffer at pH 11.7 and 298 K.
Umbrella sampling (US) is one of the methods that provide binding free energy along a physically realizable transition path—reaction coordinate, such as the distance between protein and ligand [16, 17]. In the method the relevant range of macrostates is divided into overlapping windows which are sampled according to a non-Boltzmann weighting function. The obtained biased probability distributions accumulated in these sampling windows are then combined and unbiased via statistical analysis methods such as weighted histogram analysis method (WHAM) [18] and umbrella integration, to yield the associated potential of mean force (PMF) [18]. Proper conformational sampling along the reaction coordinate is the key for an accurate estimate of the PMF, which can be improved by enhanced sampling methods like self-guided Langevin dynamics (SGLD) [19,20,21,22] or replica exchange umbrella sampling (REUS) [23, 24].
The conformational sampling accuracy can be estimated by “forward (from bound to unbound)” and “backward (unbound to bound)” USs. Ideally, at each reaction coordinate window US need proper sampling of the equilibrated conformational distribution. Due to the cost limit, the sampling at each window is affected by the previous window, which cause the “forward” US different from the “backward” US. Proper equilibrated sampling would produce little difference between them. Using enhanced sampling methods can accelerate the convergence of sampling so that accurate PMF can be obtained. For example, the REUS algorithm enables to sample various structures between the bound and the unbound states in a series of parallelized simulations by exchanging adjacent umbrella potentials.
In this paper, we will discuss our approaches to calculate the absolute binding free energies of the TEMOA host and the eight guests including our submitted results to the SAMPL6 blind challenge. Results from a similar approach applied to the CB8 host can be found in a companion paper by Han et al. (receipt ID 3z83m) [25]. By way of outline, our two FES protocols: the US with the weighted histogram analysis method (US-WHAM) and the double decoupling method (DDM) with Hamiltonian replica-exchange method and the Bennett acceptance ratio (HREM-BAR) are presented in “Materials and methods” section. Results and discussion are presented in “Results and discussion” section. We then conclude the study with our findings and future directions in “Conclusion” section.
Materials and methods
The protocol from creating the binding poses to calculating the binding free energies is depicted in Fig. 1. We generated binding pose structures in vacuum by using GalaxyDock-HG and performed equilibration MD to obtain an initial structure for free energy simulations (FES). We then calculated the binding free energies by using two schemes: US and weighted histogram analysis method (US-WHAM) scheme and double decoupling method (DDM) with the Hamiltonian replica-exchange method post-processed with the Bennett acceptance ratio (HREM-BAR) scheme (Fig. 2).
Binding poses
We first docked the host and the guest molecule through GalaxyDock-HG, a docking program which we developed specifically for the SAMPL binding free energy prediction challenge. GalaxyDock-HG finds the guest binding poses through global optimization by using the conformational space annealing (CSA) algorithm [26, 27] with the AutoDock4 scoring function [28,29,30]. GalaxyDock-HG was developed based on the Galaxy-Dock docking program [29] which is developed for protein–ligand docking. In the GalaxyDock-HG, the energy is evaluated in the continuous space, and the initial set of conformations for CSA (the initial bank) is generated by randomly perturbing the initial structures. In GalaxyDock-HG program, the following AutoDock4 scoring function is used:
where \({A_{ij}}\) and \({B_{ij}}\) are parameters for the van der Waals energy, \({C_{ij}}\) and \({D_{ij}}\) are the parameters for the hydrogen bond energy, \(h\left( {{t_{ij}}} \right)\) is the weight factor to describe hydrogen bond directionality, \({q_i}\) and \({q_j}\) are the partial charges, \(\varepsilon \left( {{r_{ij}}} \right)\) is a distance dependent dielectric constant, \(S\), \(V\), and \(\sigma\) are desolvation energy parameters. Partial charge parameters were taken from the CGENFF. A total of 50 conformations were generated as the initial bank after local energy minimization, and the bank was evolved by the CSA algorithm. It was difficult for TEMOA host–guest systems because of the steric hindrances of four methyl groups. In most docking trials, the energy minimum structures of the program were that guest molecules were inside the pocket of host molecules. However some of the minimum structures were incorrect i.e. the guest molecules were outside the binding site of the host molecules. Therefore, we continued the trial to dock until we obtained a structure in which the guest molecule is correctly inside the pocket of the host molecule. Finally, we performed around 10 times trials for TEMOA-G3 and TEMOA-G5 systems. In this way, we used the structure which has the minimum energy in the trials that finally succeeded to dock for the following simulations.
Parameters for the host and the guests were obtained by the CHARMM General Force Field (CGENFF) for organic molecules [31]. The host molecule had a net charge of − 8 due to the presence of eight carboxylate groups and the high experimental pH (11.7). Moreover, all eight guest molecules (G0–G7) contained carboxylate groups and had a charge of − 1.
All the steps described below were performed by the CHARMM [31] version c41b1 with CHARMM 36 force field [32]. Since we need not only host–guest complex systems but also guest-only systems, whose reason is described in “Hamiltonian replica-exchange method/Bennett acceptance ratio” section, we first solvated the host–guest complex systems and the guest-only systems in TIP3P explicit solvent [33, 34] in a cubic box with edge lengths of 50 Å. We added enough Na+ ions to neutralize the systems (i.e., bring the total charge of the system to zero). We then performed energy minimization using the steepest descent algorithm [35] and the adopted basis Newton–Raphson algorithm [35] for 5000 steps and 50,000 steps, respectively, with constrained heavy atoms in both the host and the guest. We heated the systems with harmonically restrained heavy atoms for 142,500 steps to 298 K and equilibrated in NVT ensemble whose temperature is 298 K for 357,500 steps. We then performed equilibration MD for 500 ps with heavy atoms in both the host and the guest harmonically restrained with force constants of \(0.5\,{\text{kcal}}/{\text{mol}}\cdot{{\AA}^2}\) in NPT ensemble, in which the temperature and the pressure were maintained constant by the Nosé–Hoover thermostat [36, 37] and Langevin piston barostat [38], respectively. Water molecules was kept rigid with SHAKE constraint [39]. The time step was set to 1 fs for each MD simulation. For the last step, we performed long equilibration MD simulations for 100–200 ns per system to obtain the initial structure for FES.
Umbrella sampling/weighted histogram analysis method
The umbrella sampling (US) [18] is a way of biased molecular dynamics (MD) to estimate free energy along a reaction coordinate. In this method, the sampling region in the conformational space is restrained to a narrow region by adding a bias potential (umbrella potential).
The bias potential can have any functional form but harmonic potentials are often used for their simplicity.
By focus on this narrow region, MD simulations can efficiently sample the conformational space and produce relative free energies at the reaction coordinates within this region. The complete free energies between the interest states can be obtained by a series of US simulations covering the whole range of reaction coordinates. For ligand binding, this method can be used not only to obtain binding free energy, but also to predict binding pose. By US ligand at different distances from the binding pocket, the binding pose can be identified as the conformation with the lowest free energy. Also, by US simulations against various cavities on protein surface, one can also identify the binding pocket according to the lowest binding free energy.
In our simulations, we selected the distance between centers of mass of the host and the guest molecules as a reaction coordinate, and slightly changed the center position of the umbrella potential with keeping the force constant same value. Although there are many possible reaction coordinates that could be used, we chose the distance as a reasonable first choice, due to its simplicity and its generality for all guests. We totally performed 40 US MD simulations. The setting of the center position of the first US simulation is set to the distance between centers of mass of the host and the guest molecules. We carefully moved the center position when the host and the guest molecules are close to each other, and gradually increased the interval between the next window and the window. The interval settings are as follows (in Å): 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.3, 0.3, 0.3, 0.3, 0.3, 0.3, 0.3, 0.3, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, and 0.5 (totally 39 intervals). Therefore, the final center position is 11.9 Å away from the initial position. Since there is a possibility that the free energy minimum is at negative values pressing the guest further into the host, we performed US simulations toward negative direction (inside the host), and found only free energy increases (data not shown). After performing the US MD simulations, the windows are combined by methods like the weighted histogram analysis method (WHAM) [18] or multistate Bennett acceptance ratio (MBAR) [40]. We used WHAM program which is developed by Grossfield laboratory version 2.0.9.1 [41] for reweighting the US results and obtained the free energy cost to pull the guest molecule from the binding pocket to outside the host molecule (ΔGpull). We estimated the error of the PMF from each US simulation by using the bootstrap error analysis with the WHAM. The error of \({{{\Delta}}}{G_{{\text{pull}}}}\) is estimated by standard error of ten independent simulations. \(- \;{{{\Delta}}}{G_{{\text{pull}}}}\) almost corresponds to the binding free energy but there are two free energy costs to be corrected (see Fig. 3): one of them is the free energy cost to give the US potential to the first window (\(\Delta G_{{{\text{rest-on}}}}\)) and the other one is the free energy cost to keep the guest molecule at the certain distance in the last window (\({{{\Delta}}}{G_{{\text{VC}}}}\)) which can be called the volume correction (VC).
Scheme of free energy calculation using umbrella sampling method. The grey box-shaped container and the black wire-shaped material represent the host and the guest molecules, respectively, and the red spring-shaped material represents the restraint (umbrella potential) which is given between the host and the guest molecules
We estimated the free energy cost for turning on the restraint (umbrella potential) by using thermodynamic integration (TI). In TI simulations, we move a mixing factor \({{{\uplambda}}}\) which is combined with each state’s potential function from the initial (λ = 0) state to the final (\({{{\uplambda}}}=1\)) state. We used 21 \({{{\uplambda}}}\) points for estimating each free energy difference associated with turning on the restraint. Simulations for TI were run in an NVT ensemble. For each \({{{\uplambda}}}\) value, we performed an equilibration for 50 ps and a production increment for 450 ps.
The volume correction term \({{{\Delta}}}{G_{{\text{VC}}}}\) is calculated by:
where \({{\text{V}}_0}\) is the standard state volume for ideal gas (1,649.76 Å3), \({k_{\text{B}}}\) is the Boltzmann constant, \(T\) is the temperature of the system, and \({V_{{\text{eff}}}}\) is the accessible volume of the guest molecule in the last window which we estimated by:
Here, we defined \({r_{{\text{max}}}}\) and \({r_{{\text{min}}}}\) as the maximum value and the minimum value of the center 95% distribution of the distance between the centers of mass of the host and the guest in the last window, respectively. Then, we can finally calculate the absolute binding free energy \({{{\Delta}}}{G_{{\text{bind}}}}\) by following equation:
The volume correction is a simple free energy estimate based on changing concentration of the guest. It could be calculated by very long simulations to a volume of 1649.76 Å3, but the simple analytic solution used here would prove to be more accurate.
Hamiltonian replica-exchange method/Bennett acceptance ratio
The double decoupling method (DDM) is a so-called “alchemical” method [42,43,44], and the scheme is represented in Fig. 4. The basic idea of this scheme is to calculate the binding free energy by taking difference between the free energy cost to eliminate the guest in the solvent and the free energy cost to eliminate the guest from the host–guest bound complex in the solvent. There are two intermolecular interactions: electrostatic interactions and van der Waals interactions in the force field which we used, therefore we divided the eliminating free energy into \({{{\Delta}}}{G_{{\text{elec-off}}}}\) and \({{{\Delta}}}{G_{{\text{vdw-off}}}}\). Because we give a restraint between the host and the guest so that the guest molecule can keep the position around the binding site even when the intermolecular interaction of the guest becomes weaker or zero, we need two correction terms: free energy cost to turn on the restraint in the complex \(\left( {{{{\Delta}}}G_{{{\text{rest-on}}}}^{{\text{C}}}} \right)\) and the free energy cost to turn off the restraint for the ghost guest which has no interactions \(\left( {{{{\Delta}}}G_{{{\text{rest-off}}}}^{{\text{C}}}} \right)\). We finally calculated the binding free energy by the thermodynamic cycle as follows:
Scheme of free energy calculation for double decoupling method with Hamiltonian replica-exchange method. The grey box-shaped container and the black wire-shaped material represent the host and the guest molecules, respectively, and the red spring-shaped material represents the restraint which is given between the host and the guest molecules. The center thinner wire-shaped material (than the left side one) represents the guest state in which the electrostatic interaction of the guest molecule is turned off. The right dotted wire-shaped material with thinner color represents the guest state in which both the electrostatic and van der Waals interactions of the guest molecule are turned off. Each free energy \({{{\Delta}}}G\) in the figure represents the free energy which is required to transition from the state of the starting point to the state of the end point of each corresponding arrow. The suffixes C and G mean the host–guest complex system and guest-only system, respectively
For the restraints to maintain the binding site pose, we used one distance restraint, two angle restraints, and three dihedral restraints which are dependent each other. We automatically picked an atom in the host and an atom in the guest which has smallest distance between the host and the guest, and used as the distance restraint. Then we picked two atoms which is different from the atom picked already and connected to the atom from the guest and the host, and used as the angle and the dihedral restraints. These restraints not only keep the position of the guest molecule, but also restrict rotations of the guest molecule. The force constants were set to as follows: \(5\,{\text{kcal}}/{\text{mol}}\cdot{{\AA}^2}\), \(20\,{\text{kcal}}/{\text{mol}}\cdot{\text{rad}}^2\), and \(20\,{\text{kcal}}/{\text{mol}}\cdot{\text{rad}}^2\) for distance, angle, and dihedral geometrical harmonic restraints, respectively. The free energy cost to turn the restraints between the guest and the host off was calculated analytically as follows [42]:
where \({k_{\text{B}}}\) is the Boltzmann constant, \(T\) is the simulation temperature, \(V\) is the volume of the simulation box, \({K_r}\) is the force constant of distance restraint, \({K_{{\theta _A}}}\) and \({K_{{\theta _B}}}\) are the force constants of angle restraints, \({K_{{\phi _A}}}\), \({K_{{\phi _B}}}\), and \({K_{{\phi _C}}}\) are the force constants of dihedral restraints, \(r\) is the distance between selected atoms in the host and the guest of the initial snapshot for FES, \({\theta _A}\) and \({\theta _B}\) are the selected angles of the initial snapshot for FES.
We used Hamiltonian replica-exchange method (HREM) [45,46,47] post-processed with the Bennett acceptance ratio (BAR) [48, 49] (hereinafter, this scheme is called HREM-BAR [11, 50,51,52]) to calculate the free energy value for turning off the intermolecular interactions, and thermodynamic integration (TI) [32] to calculate the free energy value for turning on the restraints. Although it is also possible to calculate \({{{\Delta}}}{G_{{\text{elec-off}}}}\) and \({{{\Delta}}}{G_{{\text{vdw-off}}}}\) separately, we combined those into a HREM simulation to enhance sampling. We used 11 \({{{\uplambda}}}\) points and 22 \({{{\uplambda}}}\) points for estimating \({{{\Delta}}}{G_{{\text{elec-off}}}}\) and \({{{\Delta}}}{G_{{\text{vdw-off}}}}\), respectively in the HREM simulation. Each HREM simulation was run for 1 ns, with a total of 32 ns for each system. In the TI simulations, we used 20 \({{{\uplambda}}}\) points for estimating each free energy difference associated with turning on the restraints. Simulations for TI were run in an NVT ensemble. For each \({{{\uplambda}}}\) value, we performed an equilibration for 50 ps and a production increment for 450 ps. All FES used the particle mesh Ewald method and 14 \({\AA}\) cutoffs. The time step of MD simulation was set to 1 fs.
Since the whole scheme is completely consistent with our previous SAMPL5 challenge, more details can be referred in the paper by Tofoleanu et al. [9].
Results and discussion
US-WHAM scheme example
Here, we show an example of the result of US-WHAM free energy calculation on TEMOA-G0 system. Figure 5 shows the distance between centers of mass of the host and the guest molecules. The orange line suggests the schedule for the center position of the umbrella potential. The blue line shows the actual distance between centers of mass of the host and the guest and the actual distance is fluctuated around the setting of the distance (the orange line). Figure 6 shows the distribution for the distance between the centers of mass of the host and the guest. Each color distribution suggests each independent simulation with different umbrella potential. Therefore, the distribution of the actual value of the blue line in Fig. 5 corresponds to the data shown in Fig. 6. There is sufficient overlap in the distributions of any adjacent combinations so that we can calculate the free energy difference by using a reweighting method.
Distance between centers of mass of the host and the guest on a series of umbrella sampling simulations to calculate \({{{\Delta}}}{G_{{\text{pull}}}}\) for TEMOA-G0 system. The orange line indicates the setting of the distance for each umbrella sampling simulation and the blue line indicates the actual distance value between the centers of mass
Figure 7 shows the PMF along with the reaction coordinate (the distance between centers of mass of the host and the guest). Since we performed 10 independent US simulations, there are 10 independent curves (Run1 to Run10). Each plotted point is plotted every 0.5 Å which is not necessarily coincident with the setting of the reaction coordinate. We display the error bars for the PMF of each independent simulation by using the bootstrap error analysis (however the error bars are small so that it is hard to see by eyes). We estimated the free energy cost to pull out the guest from the binding pose as the difference of the height between the last point and the reference point. The average points at each reaction coordinate is evaluated by averaging the PMF values of 10 independent simulations at the reaction coordinate. Here the error bar at the reaction coordinate is estimated by the standard error of the 10 data. Finally, the \({{{\Delta}}}{G_{{\text{pull}}}}\) value corresponds to the height of the average value (final black point). In the case of TEMOA-G0 system, \({{{\Delta}}}{G_{{\text{pull}}}}\) is 5.19 ± 0.61 kcal/mol.
FES results by US-WHAM scheme
We present each free energy term for US-WHAM scheme in Table 1 and a figure for the correlation between computed and experimental absolute free energy values in Fig. 8. We calculated the pulling and the volume correction free energy values by averaging ten independent simulations which has same initial structures but different initial velocities. Similarly, the restraint-on free energy is calculated by averaging three independent trials. Each error bar is the standard error by the FES trials.
FES results by DDM with HREM-BAR scheme
We present each free energy term for HREM-BAR scheme in Table 2 and a figure for the correlation between computed and experimental absolute free energy values in Fig. 9. The free energies for turning off the electrostatic and the van der Waals of host–guest complex systems and guest systems were calculated by averaging the results of three independent HREM simulations. The error bar of each term is the standard error of the independent FES trials, and the error bar of each complex system is calculated by using the general error propagation equation. There are no error bar for the restraint-off term of each system which is calculated analytically, because we used same initial structure and same restraint to keep the guest molecule bound to the host molecule for each complex system.
As described in “Hamiltonian replica-exchange method/Bennett acceptance ratio” section, free energy values for turning off the electrostatic and van der Waals interactions for both complex and guest systems were calculated in the same HREM simulation because we combined those steps.
Comparison between two FES methods and the experimental results
We suggest three sets of the binding free energy calculations: US-WHAM without corrections \(\left( {{{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}1}}} \right)\), US-WHAM with corrections \(\left( {{{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}} \right)\), and HREM-BAR \(\left( {{{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}} \right)\) in Table 3. We submitted the \(- {{{\Delta}}}{G_{{\text{pull}}}}\) value at the submission of the SAMPL6 competition assuming that the restraint-on term and the volume correction term approximately cancel out each other and the data are represented as \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}1}}\). The left column \(\left( {{{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}1}}} \right)\) in Table 3 corresponds to the result which we submitted for the SAMPL6 competition. Our result marked third RMSD value (1.33 kcal/mol) and top MAD value (1.02 kcal/mol) among all 45 submissions. After the submission, we performed the corrections for the restraint-on \({{{\Delta}}}{G_{{\text{rest-on}}}}\) and the volume correction \({{{\Delta}}}{G_{{\text{VC}}}}\), and evaluated \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\). The error bars were calculated by applying the error propagation equation for summation to the error of each free energy term. For each term, we tried 10 times to calculate \({{{\Delta}}}{G_{{\text{pull}}}}\) and \({{{\Delta}}}{G_{{\text{VC}}}}\), and five times to calculate \({{{\Delta}}}{G_{{\text{rest-on}}}}\).
The resulting metrics are presented in the row 10–16 in Table 3. We calculated three kinds of deviations: the root mean square deviation (RMSD), the mean absolute deviation (MAD), and the mean signed deviation (MSD) of comparison with experimental values \(\left( {{{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{exp}}}}} \right)\) as shown in the rightmost column of Table 3. Moreover, we analyzed the correlation between computed and experimental results. We represented Pearson’s coefficient (r), Kendall rank coefficient (τ), the coefficient of determination (R2), and the slope of the approximation line (m) in rows 10–16 of Table 3.
Although our results of deviations for US scheme are in the top three submissions, the correlation results were not enough reasonable. It is assumed to be derived from the fact that the calculation precision of US scheme is low and the error bars are large. On the other hand, although the results for the HREM-BAR has larger deviations than the US-WHAM results, the correlations for the HREM-BAR show better results. We also calculated the deviation and the correlation values between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) values. Because we use the same force field, the same program package, and the same initial conformation for the two schemes, those two results should ideally agree, and the agreement between the results from different computational schemes is more important than the agreement between each computational result and experimental result. Although the deviations RMSD and MAD between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) are smaller than the deviations between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{exp}}}}\), the deviations between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) are larger than the deviations between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{exp}}}}\). However, the correlation results between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) (0.97, 0.79, 0.94) (Pearson’s coefficient (r), Kendall rank coefficient (τ), and the coefficient of determination (R2), respectively) are larger than the correlation results between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{exp}}}}\) (0.70, 0.36, 0.49) and the correlation results between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{exp}}}}\) (0.74, 0.43, 0.54), indicating our two schemes US-WHAM and HREM-BAR are strongly correlated. Here, the fact that the values of MSD and MAD between \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) and \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) agree indicates that all the respective \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{HB}}}}\) values are larger than the \({{{\Delta}}}G_{{{\text{bind}}}}^{{{\text{US}}2}}\) values. In our DDM scheme which is an alchemical method, the intermolecular interactions of the guest molecule are turned off and completely eliminated. However, since our simulations systems are cubes whose edge length are around 50 Å, it is difficult to ignore the contribution of the volume change that the guest molecule disappears and appears. If such problems are solved, HREM-BAR scheme is expected to provide better results than US-WHAM scheme because the correlation results of the HREM-BAR are better (Table 4).
Conclusion
The RMSD and the MAD results for the TEMOA-guest systems of our US-WHAM scheme were relatively low and ranked third and top, respectively, among all submissions in the SAMPL6 competition. However, its convergence still has problems. Our present US-WHAM scheme is just an “one-way” unbinding simulation, meaning to extract the guest molecule from the binding pocket of the host molecule. Therefore, the absolute binding free energy calculated by the scheme largely depends on the initial structure. In order to improve those tendency, a way to repeat “round-trip” unbinding US and binding US simulations several times is considered. Alternatively, it would be more efficient to create multiple bound state using the docking algorithm and perform FES. In addition, the replica-exchange US (REUS) [23, 24] can be considered to be a better method in this regard. We attempted to calculate binding free energies using the REUS, however the simulations are failed because the guest molecule crashed to the side face or the entrance of the host molecule and did not return to the binding pocket during the rebinding process. This is because the reaction coordinate is set to merely the distance between the centers of mass of the host and guest molecules, allowing the guest molecule to move around the host molecule. This behavior is considered to lead inaccuracy results and insufficient sampling. In order to overcome such difficulties, the method needs an additional cylinder-like restraint along with the symmetry axis of the host molecule to the guest molecule [16, 17]. Our next direction for US-WHAM scheme is to calculate binding free energies by using the REUS and/or its applied method.
We also calculated the binding free energy by using HREM-BAR scheme to compare with the US-WHAM scheme. Although the deviation between the two schemes are large, correlations between the two schemes are high. It is suggested that the TI calculation has less accuracy than HREM-BAR [9], therefore we are attempting to apply the HREM-BAR to the restraint-on step. Moreover, various kinds of constraints can be considered when computing the complex free energies. It is also a worth challenge to try various restraints. Another restraint may also be possible to facilitate sampling various structures of the bound state without relying heavily on the initial conformation of the FES.
Despite that our FES depends on the initial conformations, we merely chose the final conformations of the equilibrium simulation, the results are encouraging. A more rigorous screening method to determine the initial conformations should be established. Simply, a process such as creating a free energy landscape of the equilibrium simulation structures and picking up a structure (or some structures) which have minimum free energy, is conceivable. In addition, it may be effective to use better sampling methods such as self-guided Langevin dynamics (SGLD) to sample bound states more efficiently.
References
Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303(5665):1813–1818
De Vivo M, Masetti M, Bottegoni G, Cavalli A (2016) Role of molecular dynamics and related methods in drug discovery. J Med Chem 59(9):4035–4061
Guthrie JP (2009) A blind challenge for computational solvation free energies: introduction and overview. J Phys Chem 113:4501–4507
Gallicchio E, Chen H, Chen H et al (2015) BEDAM binding free energy predictions for the SAMPL4 octa-acid host challenge. J Comput Aided Mol Des 29(4):315–325
Ellingson BA, Geballe MT, Wlodek S, Bayly CI, Skillman AG, Nicholls A (2014) Efficient calculation of SAMPL4 hydration free energies using OMEGA, SZYBKI, QUACPAC, and Zap TK. J Comput Aided Mol Des 28(3):289–298
Nicholls A, Mobley DL, Guthrie JP et al (2008) Predicting small-molecule solvation free energies: an informal blind test for computational chemistry. J Med Chem 51(4):769–779
Beckstein O, Fourrier A, Iorga BI (2014) Prediction of hydration free energies for the SAMPL4 diverse set of compounds using molecular dynamics simulations with the OPLS-AA force field. J Comput Aided Mol Des 28(3):265–276
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24(4):259–279
Tofoleanu F, Lee J, Pickard IVFC et al (2017) Absolute binding free energies for octa-acids and guests in SAMPL5. J Comput Aided Mol Des 31(1):107–118
Lee J, Tofoleanu F, Pickard FC et al (2017) Absolute binding free energy calculations of CBClip host–guest systems in the SAMPL5 blind challenge. J Comput Aided Mol Des 31(1):71–85
König G, Brooks BR (2012) Predicting binding affinities of host–guest systems in the SAMPL3 blind challenge: the performance of relative free energy calculations. J Comput Aided Mol Des 26(5):543–550
Gan H, Benjamin CJ, Gibb BC (2011) Nonmonotonic assembly of a deep-cavity cavitand. J Am Chem Soc 133(13):4770–4773
Gibb CLD, Gibb BC (2014) Binding of cyclic carboxylates to octa-acid deep-cavity cavitand. J Comput Aided Mol Des 28(4):319–325
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) The SAMPL4 host–guest blind prediction challenge: an overview. J Comput Aided Mol Des 28(4):305–317
Sullivan MR, Sokkalingam P, Nguyen T, Donahue JP, Gibb BC (2017) Binding of carboxylate and trimethylammonium salts to octa-acid and TEMOA deep-cavity cavitands. J Comput Aided Mol Des 31(1):21–28
Woo H-J, Roux B (2005) Calculation of absolute protein–ligand binding free energy from computer simulations. Proc Natl Acad Sci 102(19):6825–6830
Gumbart JC, Roux B, Chipot C (2013) Efficient determination of protein–protein standard binding free energies from first principles. J Chem Theory Comput 9(8):3789–3798
Roux B (1995) The calculation of the potential of mean force using computer simulations. Comput Phys Commun 91(1–3):275–282. https://doi.org/10.1016/0010-4655(95)00053-I
Wu X, Damjanovic A, Brooks BR (2012) Efficient and unbiased sampling of biomolecular systems in the canonical ensemble: a review of self-guided Langevin dynamics. Adv Chem Phys 150:255
Wu X, Brooks BR (2003) Self-guided Langevin dynamics simulation method. Chem Phys Lett 381(3–4):512–518
Wu X, Brooks BR, Vanden-Eijnden E (2016) Self-guided Langevin dynamics via generalized Langevin equation. J Comput Chem 37(6):595–601
Wu X, Brooks BR (2011) Toward canonical ensemble distribution from self-guided Langevin dynamics simulation. J Chem Phys 134(13):04B605
Murata K, Sugita Y, Okamoto Y (2004) Free energy calculations for DNA base stacking by replica-exchange umbrella sampling. Chem Phys Lett 385(1–2):1–7
Sugita Y, Kitao A, Okamoto Y (2000) Multidimensional replica-exchange method for free-energy calculations. J Chem Phys 113(15):6042–6051
Han K, Hudson PS, Jones MR, Nishikawa N, Tofoleanu F, Brooks BR (2018) Prediction of CB [8] host–guest binding free energies in SAMPL6 using the double-decoupling method. J Comput Aided Mol Des. https://doi.org/10.1007/s10822-018-0144-8
Shin W-H, Heo L, Lee J, Ko J, Seok C, Lee J (2011) LigDockCSA: protein–ligand docking using conformational space annealing. J Comput Chem 32(15):3226–3232
Lee J, Scheraga HA, Rackovsky S (1997) New optimization method for conformational energy calculations on polypeptides: conformational space annealing. J Comput Chem 18(9):1222–1232
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28(6):1145–1152
Shin W-H, Lee GR, Seok C (2015) Evaluation of galaxydock based on the community structure–activity resource 2013 and 2014 benchmark studies. J Chem Inf Model 56(6):988–995
Shin W-H, Kim J-K, Kim D-S, Seok C (2013) GalaxyDock2: protein–ligand docking using beta-complex and global optimization. J Comput Chem 34(30):2647–2656
Vanommeslaeghe K, Hatcher E, Acharya C et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690
Kirkwood JG (1935) Statistical mechanics of fluid mixtures. J Chem Phys 3(5):300–313
MacKerell AD Jr, Bashford D, Bellott M et al (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102(18):3586–3616
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4(2):187–217
Hoover WG (1985) Canonical dynamics: equilibrium phase-space distributions. Phys Rev A 31(3):1695
Nosé S (1984) A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys 81(1):511–519
Feller SE, Zhang Y, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103(11):4613–4621
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341
Shirts MR, Chodera JD (2008) Statistically optimal analysis of samples from multiple equilibrium states. J Chem Phys 129(12):124105. https://doi.org/10.1063/1.2978177
Grossfield A. WHAM: the weighted histogram analysis method, version 2.0.9.1, http://membrane.urmc.rochester.edu/content/wham
Boresch S, Tettinger F, Leitgeb M, Karplus M (2003) Absolute binding free energies: a quantitative approach for their calculation. J Phys Chem B 107(35):9535–9551
Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA (2007) Predicting absolute ligand binding free energies to a simple model site. J Mol Biol 371(4):1118–1134
Gilson MK, Given JA, Bush BL, McCammon JA (1997) The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys J 72(3):1047–1069
Itoh SG, Okumura H, Okamoto Y (2010) Replica-exchange method in van der Waals radius space: overcoming steric restrictions for biomolecules. J Chem Phys 132(13):134105
Itoh SG, Okumura H (2013) Hamiltonian replica-permutation method and its applications to an alanine dipeptide and amyloid-β (29–42) peptides. J Comput Chem 34(29):2493–2497
Fukunishi H, Watanabe O, Takada S (2002) On the Hamiltonian replica exchange method for efficient sampling of biomolecular systems: application to protein structure prediction. J Chem Phys 116(20):9058–9067
Bennett CH (1976) Efficient estimation of free energy differences from Monte Carlo data. J Comput Phys 22(2):245–268
Shirts MR, Bair E, Hooker G, Pande VS (2003) Equilibrium free energies from nonequilibrium measurements using maximum-likelihood methods. Phys Rev Lett 91(14):140601
König G, Pickard FC, Mei Y, Brooks BR (2014) Predicting hydration free energies with a hybrid QM/MM approach: an evaluation of implicit and explicit solvation models in SAMPL4. J Comput Aided Mol Des 28(3):245–257
König G, Hudson PS, Boresch S, Woodcock HL (2014) Multiscale free energy simulations: an efficient method for connecting classical MD simulations to QM or QM/MM free energies using non-Boltzmann Bennett reweighting schemes. J Chem Theory Comput 10(4):1406–1419
König G, Bruckner S, Boresch S (2009) Unorthodox uses of Bennett’s acceptance ratio method. J Comput Chem 30(11):1712–1718
Acknowledgements
The authors would like to thank Gerhard König, John Legato, Minkyung Baek and Chaok Seok for helpful discussion and technical assistance. This work was partially supported by the intramural research program of the National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health and employed the high-performance computational capabilities of the LoBoS and Biowulf Linux clusters at the National Institutes of Health. (http://www.lobos.nih.gov and http://www.biowulf.nih.gov.)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nishikawa, N., Han, K., Wu, X. et al. Comparison of the umbrella sampling and the double decoupling method in binding free energy predictions for SAMPL6 octa-acid host–guest challenges. J Comput Aided Mol Des 32, 1075–1086 (2018). https://doi.org/10.1007/s10822-018-0166-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0166-2