Abstract
Purpose
To investigate the feasibility of the application of conventional in vitro fertilization (cIVF) for couples undergoing preimplantation genetic testing for aneuploidies (PGT-A) with non-male factor infertility.
Methods
To evaluate the efficiency of sperm whole-genome amplification (WGA), spermatozoa were subjected to three WGA protocols: Picoplex, ChromInst, and multiple displacement amplification (MDA). In the clinical studies, 641 couples who underwent PGT-A treatment for frozen embryos between January 2016 and December 2021 were included to retrospectively compare the chromosomal and clinical outcomes of cIVF and intracytoplasmic sperm injection (ICSI). Twenty-six couples were prospectively recruited for cIVF and PGT-A treatment between April 2021 and April 2022; parental contamination was analyzed in biopsied samples; and 12 aneuploid embryos were donated to validate the PGT-A results.
Results
Sperm DNA failed to amplify under Picoplex and ChromInst conditions but could be amplified using MDA. In frozen PGT-A cycles, no significant differences in the average rates of euploid, mosaic, and aneuploid embryos per cycle between the cIVF-PGT-A and ICSI-PGT-A groups were observed. The results of the prospective study that recruited couples for cIVF-PGT-A treatment showed no paternal contamination and one case of maternal contamination in 150 biopsied trophectoderm samples. Among the 12 donated embryos with whole-chromosome aneuploidy, 11 (91.7%) presented uniform chromosomal aberrations, which were in agreement with the original biopsy results.
Conclusions
Under the Picoplex and ChromInst WGA protocols, the risk of parental contamination in the cIVF-PGT-A cycles was low. Therefore, applying cIVF to couples with non-male factor infertility who are undergoing PGT-A is feasible.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Preimplantation genetic testing (PGT) is a method for testing single-gene disorders and chromosomal and mitochondrial defects in embryos fertilized in vitro, thereby avoiding the transmission of the defects to the offspring. PGT was first introduced for human embryos in 1989 by demonstrating that it was possible to detect the sex of an embryo [1]. At that time, conventional in vitro fertilization (cIVF) was the only choice for insemination. Once the intracytoplasmic sperm injection (ICSI) technique was invented [2], it quickly became the predominant insemination method in PGT cycles based on the assumption that any risk of extraneous sperm contamination was eliminated and clinical outcomes were improved. Furthermore, ICSI was previously recommended by the American Society for Reproductive Medicine (ASRM), the European Society of Human Reproduction and Embryology (ESHRE), and the Preimplantation Genetic Diagnosis International Society (PGDIS) for PGT cycles [3,4,5]. However, scientific evidence of the benefits of ICSI is lacking.
ICSI is a “non-natural” invasive technique that circumvents natural sperm selection mechanisms, carries the risk of injecting biochemical contaminants and exogenous DNA, and may disturb the ooplasm and meiotic spindles. The long-term safety of ICSI offspring is a concern [6]. Many researchers broadly agree that the use of ICSI for non-male factor infertility does not improve clinical outcomes compared to cIVF [7,8,9,10]; however, the financial burden on patients and the workload in embryology laboratories are increased [11]. Recently, the ASRM suggested that ICSI for PGT in the absence of male factor infertility should be limited to cases in which contamination with extraneous sperm could affect the accuracy of the test results [12]. However, it is unclear under what conditions the accuracy of PGT results is disturbed by extraneous sperm.
Clinical studies have compared the prevalence of embryonic chromosomal abnormalities between patients with cIVF and ICSI during PGT cycles. Similar abnormal genetic rates and negligible paternal contamination have been observed for cIVF and ICSI in cycles of PGT for monogenic disorders [13]. In cycles of PGT for aneuploidies (PGT-A), two sibling-oocyte studies showed that blastocysts created with cIVF and ICSI had similar rates of aneuploidy and mosaicism [14, 15], whereas one retrospective cohort study identified a trend toward higher rates of mosaicism in cIVF versus ICSI [16]. Recently, a retrospective study with a large sample size demonstrated that, in the context of non-male factor infertility, ICSI resulted in an 11% lower embryo euploidy rate compared with cIVF [17]. The aforementioned findings provide evidence for the use of cIVF in PGT cycles. However, further studies are required to confirm its reliability and validity.
In this study, we examined the efficiency of sperm DNA amplification using various whole-genome amplification (WGA) protocols. A retrospective cohort study was performed to compare the chromosomal and clinical outcomes between the two insemination methods in frozen PGT-A cycles, and the PGT-A results in cIVF were validated using data from prenatal diagnoses and spontaneous abortuses. Furthermore, we prospectively recruited couples for cIVF and PGT-A treatments to evaluate parental contamination in cIVF-inseminated embryos and verified the PGT-A results using donated aneuploid embryos.
Materials and methods
Study design and patients
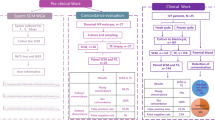
This study consisted of two parts, as illustrated in Fig. 1.
Part I: WGA and copy number variation sequencing (CNV-seq) of human sperm
First, different numbers (n = 1, 3, 5, and 10) of spermatozoa were used for three standard WGA protocols (Picoplex, ChromInst, and multiple displacement amplification (MDA)), followed by CNV-seq, and each group was replicated twice. Second, to explore the effect of lyase concentration and lysis time on sperm amplification, 10 spermatozoa were amplified using non-standard conditions of the ChromInst protocol, which combined three lyase concentrations (1* lysis enzyme; 2.5* lysis enzyme; 2.5* lysis enzyme + 0.1 mg/ml Qiagen Proteinase K) and three lysis time (75 °C, 10 min; 95 °C, 4 min; 22 °C, hold; 55 °C, 15 min; 75 °C, 10 min; 95 °C, 10 min; 22 °C, hold; 55 °C, 30 min; 75 °C, 10 min; 95 °C, 10 min; 22 °C, hold), followed by CNV-seq, with three replicates per group. Third, to mimic sperm contamination in trophectoderm (TE) biopsy, spermatozoa were mixed with five aneuploid human embryonic stem cells (hESCs) and subjected to three standard WGA protocols, followed by CNV-seq. The ratios of sperm–hESC were 1:5, 3:5, 5:5, and 10:5 and were prepared in triplicate.
Part II: comparison of embryo ploidy and clinical outcomes between cIVF and ICSI and verification of PGT-A results in cIVF
In our clinic, ICSI is routinely performed for fresh PGT cycles. Some patients with abnormal pregnancies (primarily spontaneous abortions with chromosomal abnormalities or recurrent implantation failure) in fresh cIVF or ICSI cycles choose PGT-A for their surplus frozen embryos (frozen PGT). A retrospective cohort study enrolling all patients who underwent PGT-A treatment for frozen embryos and had at least one blastocyst for biopsy between January 2016 and December 2021 was performed to compare the chromosomal and clinical outcomes between cIVF and ICSI, and the cytogenetic results of prenatal diagnoses and spontaneous abortuses were followed up. We further conducted a prospective study by recruiting couples for cIVF and PGT-A treatments (fresh cIVF-PGT) between April 2021 and April 2022. The inclusion criteria were compliance with the indications for PGT-A, normal ovarian reserve, and normal sperm quality, according to the World Health Organization criteria [18]. Parental contamination was analyzed in all biopsied TE samples from the recruited couples. To validate the concordance of the CNV-seq results between the biopsied TE samples and the remaining blastocysts, donated aneuploid blastocysts were dissected into two sections, inner cell mass (ICM) and TE, according to the method described by Capalbo et al. [19].
Ethical approval
This study was approved by the ethics committee of the Reproductive and Genetic Hospital of CITIC-Xiangya. Except for those enrolled in the retrospective study, all patients provided informed consent before any study-specific procedures were performed. For the retrospective study, which was based on an analysis of medical electronic records, informed consent was exempted by the ethics committee under the condition that the data were anonymously analyzed.
Sperm collection and mixture with hESC
Considering the structural changes in sperm after the acrosome reaction and their possible effect on the WGA results, we collected zona pellucida (ZP)-bound sperm by mechanically stripping arrested day 6 embryos. Ten patients who underwent cIVF donated the arrested embryos. The embryos were placed in 10 μl drops of G-MOPS (Vitrolife, Goteborg, Sweden) covered with paraffin oil (Ovoil, Vitrolife). ZP-bound sperm was stripped down by repeated pipetting of the embryos in and out of a 60-μm glass pipette. Based on the study design, the corresponding numbers of detached sperm were transferred with an ICSI micropipette (CooperSurgical, USA) to 2 μl drops of G-MOPS. Each drop was pipetted up and down three times and transferred to a 0.2-ml polymerase chain reaction (PCR) tube (Axygen, USA). After tubing, the drops were examined under an inverted microscope to confirm the absence of residual sperm.
In the mixing experiment, five cells from an established aneuploid hESC (46, XN, + 2p (pter → p16.2, ~ 54 Mb, × 3), − 6q (q27 → qter, ~ 4.9 Mb, × 1)) were transferred into a 0.2-ml PCR tube and mixed with the corresponding amounts of detached ZP-bound sperm. The cells were obtained from an established hESC bank at the National Engineering and Research Center of Human Stem Cells. The cell line was derived from an unbalanced translocation embryo of an individual with reciprocal translocation, and the karyotype of cell line was confirmed using fluorescence in situ hybridization and CNV shotgun sequencing as previously described [20].
Laboratory procedures
Oocyte retrieval was performed under ultrasound guidance 34–36 h after human chorionic gonadotrophin (hCG) administration. All cumulus-oocyte complexes (COCs) were placed in fertilization medium (G-IVF, Vitrolife) at 37 °C in an atmosphere of 6% CO2 for 3–5 h before inseminating by cIVF or ICSI. For patients with cIVF, 100,000 motile sperm were added to 1 ml fertilization medium in a tube (BD Falcon™ 352,003), and subsequently, 2–3 COCs were transferred into the tube. Sperm COCs were co-cultured overnight at 37 °C and 6% CO2 in an incubator (Thermo Forma 3121). For patients who underwent ICSI, the COCs were stripped using hyaluronidase (Vitrolife), and only oocytes that extruded the first polar body were microinjected. After injection, the oocytes were transferred to 20 μl fertilization medium and cultured individually overnight in the tri-gas incubator (ASTEC APM-50D) at 37 °C, 6% CO2, and 5% O2. Normal fertilization was identified 16–18 h after insemination by the presence of two pronuclei (2PN). Subsequently, all zygotes were transferred to cleavage medium (G1.5, Vitrolife). On day 3, the embryos were scored using Puissant’s criterion and then transferred, cryopreserved, or cultured to the blastocyst stage based on clinical indications and the patient’s willingness. In cycles of blastocyst culture, blastocysts that scored ≥ 4BC on day 5 according to the grading system developed by Gardner and Schoolcraft [21] were considered suitable for transfer; the remaining blastocysts (≥ 4BC) on days 5–7 were cryopreserved. Embryos were vitrified using the Kitazato vitrification kit (Kitazato Biopharma, Shizuoka, Japan) in combination with a closed high-security vitrification straw (Cryo Bio System, France).
Frozen cycles of thawed cleavage-stage embryos and thawed blastocysts were included in the frozen PGT. The thawed cleavage-stage embryos were transferred to blastocyst medium (G2.5, Vitrolife), and TE biopsy was performed on days 5–7 if the embryos reached a morphological grade of at least 4BC. The thawed blastocysts were transferred to G2.5 medium and cultured for 1–4 h. Re-expanded blastocysts were considered surviving and suitable for biopsy. A small breach (approximately 10 μm) was created in the ZP by one or two laser pulses immediately before biopsy, and approximately 3–8 TE cells were aspirated into the biopsy pipette and separated by laser-mediated drilling [22], and sperm contamination was carefully avoided under an inverted microscope. Blastocysts were vitrified within 1–2 h after TE biopsy, and the biopsied TE cells were subjected to next-generation sequencing. Only euploid embryos were selected for transfer in the frozen embryo transfer cycles; mosaic embryos were not considered. No more than two blastocysts were transferred to each patient, and a single embryo transfer was recommended.
In fresh cIVF-PGT, cIVF-inseminated embryos were cultured to the blastocyst stage, and the blastocysts were biopsied and cryopreserved as described above.
WGA protocols and sequencing
Three different strategies were used to perform WGA according to the manufacturer’s instructions: Picoplex (Rubicon Genomics, USA), ChromInst (Yikon Genomics, China), and MDA (ChromLong, Yikon Genomics, China). Picoplex and ChromInst were used for the clinically biopsied TE samples. The DNA concentration of the purified library mixture was determined by Qubit®2.0 (Thermo Fisher Scientific, USA).
To analyze the ploidy status of sperm cells, mixed samples, and biopsied TE samples, the library was sequenced using a NextSeq 550 sequencer (Illumina, Inc., USA) with a single-ended read length of 55 bp. Approximately two million (sperm cells and mixed samples) or ten million (biopsied TE samples) raw reads were generated for each sample. Whole-chromosome aneuploidies, mosaicism, and segmental aneuploidies (deletion or duplication > 4 Mb) were identified in biopsied TE samples. Embryos were defined as mosaics if the equivalent of 30–70% of the cells were abnormal. Less than 30% of abnormal cells were classified as euploid, and over 70% of abnormal cells were classified as aneuploid. The CNV-seq platform was previously validated for mosaicism detection using cell mixing experiments [23].
Genotyping assay of samples and parental genomes
The genotypes of each TE and parental genome were detected using an Infinium Asian Screening Array bead chip (Illumina Inc., USA). Signal scanning was performed using the iScan system (Illumina Inc., USA). Genomic DNA was extracted from parental peripheral blood. Genomic DNA and amplified DNA from the TE samples were linearly amplified, fragmented, precipitated, and hybridized according to the manufacturer’s instructions. The genotype of each sample was analyzed using GenomeStudio software (version 2.0, Illumina, Inc., USA), and the B allele frequency and log R ratio values of all single nucleotide polymorphisms were generated simultaneously. The risk of parental DNA contamination in samples can be effectively detected using a quantification parental contamination testing (qPCT) model based on allelic ratio analysis that uses the sequencing results of WGA products [24].
Outcome measures
In the retrospective study, only the first embryo transfer cycle after PGT-A treatment was used to analyze pregnancy outcomes. Clinical pregnancy was diagnosed by ultrasound detection of a gestational sac 2 weeks after a positive hCG test. Early miscarriage was defined as the spontaneous loss of intrauterine pregnancy before 12 weeks of gestation. Ongoing pregnancy was defined as the presence of a fetal heartbeat on ultrasonography at 12 weeks of gestation.
Statistical analysis
Continuous variables were described as mean ± standard deviation or median (quartile) according to distribution. A two-sample t-test was used for normally distributed values, and the Mann–Whitney U test was used for skewed data. Categorical variables were described as frequencies or percentages and analyzed using Pearson’s chi-square test or Fisher’s exact test, as appropriate. All statistical analyses were conducted using IBM SPSS Statistics 25, and a P value < 0.05 was considered statistically significant.
In the retrospective design of frozen PGT-A cycles, euploidy, aneuploidy, and mosaicism rates were calculated per cycle and averaged for the cIVF and ICSI groups to compare chromosomal outcomes. Linear regression analysis was conducted to correct for potential confounding factors (female age, body mass index (BMI), male age, sperm source, sperm concentration, sperm motility, number of oocytes retrieved, developmental stage of embryos at freezing in their corresponding fresh cycles, indications for PGT-A, and percentage of good-quality blastocysts in frozen PGT cycles). To compare pregnancy outcomes in the first post-PGT embryo transfer cycle, logistic regression analysis was performed to adjust for confounders (female age, BMI, sperm source, sperm concentration, sperm motility, number of oocytes retrieved, developmental stage of embryos at freezing in their corresponding fresh cycles, indications for PGT-A in frozen cycles, number of embryos transferred, transfer of good-quality blastocysts, protocol for endometrial preparation, and endometrial thickness in embryo transfer cycles).
Results
Whole-genome amplification of sperm and sperm–hESC mixture
The WGA experiment, in which one, three, five, or ten spermatozoa were collected, exhibited no DNA smears for the Picoplex and ChromInst products, whereas positive results were observed for the MDA products. After CNV-seq, the Picoplex and ChromInst products were unable to generate qualified results, whereas high-quality CNV-seq results were obtained from the MDA products of the 10 spermatozoa (Fig. 2 and Supplementary Table S1). ChromInst-WGA of 10 spermatozoa with high lyase concentrations and prolonged lysis time failed to amplify sperm DNA and generate qualified CNV-seq results (Supplementary Table S1).
WGA and sequencing results of sperm. A Sperm cells (right picture) detached from zona pellucida of an arrested embryo (left picture). B Electrophoresis of the WGA products from 1, 3, 5, and 10 sperm cells amplified by Picoplex, ChromInst, and MDA. P, positive control; N, negative control. C CNV-seq results of WGA products amplified from 10 sperm cells using Picoplex, ChromInst, and MDA. D CNV-seq results of WGA products from 10 sperm cell and 5 hESC mixtures amplified by Picoplex, ChromInst, and MDA
All 36 sperm–hESC samples were successfully amplified, and CNV-seq results were obtained. The results showed that, when one, three, or five spermatozoa were added to five aneuploid hESCs, no change in the CNV-seq results of hESCs was observed, regardless of the WGA protocol (Supplementary Table S1). When 10 spermatozoa were added, the CNV-seq results of hESCs remained unchanged using Picoplex and ChromInst but changed from aneuploidy to mosaicism using MDA (Fig. 2 and Supplementary Table S1).
Embryo ploidy and pregnancy outcomes between cIVF-PGT-A and ICSI-PGT-A
In frozen PGT, 1390 blastocysts from 442 cIVF cycles and 536 blastocysts from 199 ICSI cycles were biopsied. Among them, 1387 and 536 blastocysts had conclusive chromosomal diagnoses in the cIVF and ICSI groups, respectively. The numbers of euploid, mosaic, and aneuploid embryos were 757, 173, and 457 in the cIVF group and 293, 77, and 166 in the ICSI group, respectively.
The baseline characteristics of the two groups are displayed in Supplementary Table S2. No significant differences were observed in the average rates of euploid, mosaic, and aneuploid embryos per cycle between cIVF-PGT-A and ICSI-PGT-A (50.0 (27.1, 75.0) versus 50.0 (25.0, 77.5), P = 0.990; 0 (0, 25.0) versus 0 (0, 25.0), P = 0.766; 25.0 (0, 57.5) versus 25.0 (0, 50.0), P = 0.286, respectively). Among the first embryo transfer cycles post-PGT, clinical pregnancy, early miscarriage, and ongoing pregnancy rates were not significantly different between the cIVF-PGT-A and ICSI-PGT-A groups (59.4% versus 55.6%, P = 0.148; 9.6% versus 12.0%, P = 0.792; 53.0% versus 48.1%, P = 0.205, respectively) (Table 1).
Parental contamination analysis of TE samples in fresh cIVF-PGT
Twenty-six patients were recruited in fresh cIVF-PGT, with an average age of 35.0 ± 4.0 years, a BMI of 20.7 ± 2.4 kg/m2, and AMH levels of 5.3 ± 4.0 ng/ml. In total, 432 oocytes were obtained and produced 285 2PN zygotes, 276 cleaved embryos, and 153 available blastocysts. After CNV-seq, 150 blastocysts had a conclusive chromosomal diagnosis, and the rates of euploidy, mosaicism, and aneuploidy were 52.0%, 11.3%, and 36.7%, respectively.
Based on the qPCT model, parental contamination was analyzed in the biopsied TE samples of all 150 blastocysts. No paternal contamination was detected in any sample. Maternal contamination was observed in one TE sample at a contamination level of 10% (Table 2).
Validating the PGT-A results in cIVF-PGT
To validate the PGT-A results of the biopsied TE sample in the remaining embryo, 12 blastocysts with whole-chromosome aneuploidy donated by eight patients in fresh cIVF-PGT were dissected into two sections: ICM and TE. In 11 embryos, the CNV-seq results of the dissected ICM and TE samples were consistent with the PGT-A results. One embryo displayed identical CNV-seq results in the clinically biopsied TE and dissected ICM samples, but an extra segmental mosaicism was identified in the dissected TE sample (Table 2).
To explore whether there were false-negative diagnoses in cIVF-PGT-A, the cytogenetic results of the prenatal diagnoses and abortion samples were analyzed. We tracked 32 prenatal diagnoses (one from fresh PGT and 31 from frozen PGT) and eight cytogenetic results from abortion samples (all from frozen PGT), all of which demonstrated normal karyotypes.
After cIVF-PGT-A administration, 173 infants (10 from fresh PGT and 163 from frozen PGT) were born. The sex of all the infants was consistent with the PGT-A results.
Discussion
Our study showed that under the conditions of Picoplex and ChromInst, sperm DNA failed to amplify, even at high lyase concentrations and prolonged lysis time. Moreover, the artificial mixing of hESCs with spermatozoa did not alter the CNV-seq results of hESCs. In contrast, sperm DNA was amplified, and the CNV-seq results of hESCs shifted from aneuploidy to mosaicism in the mixing experiment using MDA. Similar results have been recently reported [14, 24, 25]. Amplification of sperm DNA requires strong cell lysis conditions, which typically involve incubation with dithiothreitol and alkalis to induce sperm chromatin decondensation [26,27,28]. The lysis conditions for Picoplex and ChromInst were milder, which effectively amplify the biopsied embryo samples without amplifying sperm DNA. The alkaline lysis used in MDA is more aggressive for cell lysis and thus achieves sperm DNA amplification, indicating a high risk of paternal contamination in MDA-based cIVF-PGT.
A recent meta-analysis suggested that, compared with cIVF, there is no indication of an increased risk of chromosomal abnormalities in ICSI offspring [29]. However, data on the ploidy of preimplantation embryos in cIVF and ICSI are limited. Our results showed that under aggressive conditions of cell lysis before WGA, the CNV-seq results of hESCs changed from aneuploidy to mosaicism upon mixing with spermatozoa, and we speculated that the mosaicism rate would increase in cIVF-PGT-A cycles if sperm contamination could disturb the PGT-A results. Our study identified no difference in the mosaicism rate between the two insemination methods in frozen PGT cycles, suggesting that cIVF is unlikely to significantly affect the CNV-seq results of embryos. Deng et al. observed similar mosaicism rates for cIVF and ICSI [15]. A study reported that cIVF increased the mosaicism rate compared to ICSI; however, no significant difference was observed between the two groups [16]. In our opinion, cIVF can be safely applied in PGT-A cycles because visual inspection of spermatozoa under an inverted microscope largely avoids the presence of spermatozoa in biopsied TE pieces, and sperm DNA cannot be amplified under the WGA conditions used for biopsied TE samples.
Research on the clinical effectiveness of cIVF in patients meeting the indications for PGT-A is limited. For advanced age patients with non-male factor infertility, studies have demonstrated that no advantage for ICSI over cIVF in terms of the rates of fertilization, top-quality embryo, clinical pregnancy, miscarriage, and live birth has been observed [30,31,32,33]. To the best of our knowledge, no previous study has compared the clinical outcomes of cIVF and ICSI in patients with a history of recurrent implantation failure or miscarriage. Our study analyzed pregnancy outcomes in frozen PGT cycles of patients who primarily experienced abortion with chromosomal aberration or recurrent implantation failure after cIVF or ICSI treatment and observed that the rates of clinical pregnancy, early miscarriage, and ongoing pregnancy were similar between the two groups of insemination methods.
A recently developed method, qPCT, was used for quantitative parental contamination testing [24]. The qPCT model can effectively detect the risk of parental DNA contamination in samples based on allelic ratio analysis using the sequencing results of the WGA products. No paternal contamination was detected in any blastocysts from fresh cIVF-PGT-A, indicating that the risk of paternal contamination was negligible in fresh cIVF-PGT-A cycles using Picoplex and ChromInst. The qPCT results for one blastocyst showed a 10% level of maternal contamination, and the contamination rate (0.67%, 1/150) was similar to that reported by Dong et al. (0.83%) [24]. The aforementioned finding suggests that maternal contamination should be carefully avoided in PGT cycles. Residual cumulus cells on ZP should be completely removed before biopsy, regardless of whether the embryos are inseminated by cIVF or ICSI. Parental contamination testing methods are required to obtain accurate PGT results. The qPCT model used in this study is sensitive; however, the requirement of parental genetic information limits its routine clinical application. Single-cell whole-genome methylation sequencing was recently developed to simultaneously assess chromosomal ploidy and epigenomic status. Different DNA methylation signatures of oocytes, sperm, and embryos can help trace the cellular origins of biopsied samples and filter out irrelevant data from parental contamination [34, 35].
To verify the cIVF-PGT-A results, we dissected 12 embryos classified as whole-chromosome aneuploids into the ICM and TE. The CNV-seq results of the clinically biopsied samples were almost identical to those of ICM and TE. This result is consistent with the observations of previous studies that demonstrated the accuracy of ICSI-PGT-A using TE biopsy for whole-chromosome aneuploidy detection [36, 37]. Additionally, we tracked prenatal diagnoses, cytogenetic results of abortion samples, and newborn sex in cIVF-PGT-A cycles and found no misdiagnoses. These pieces of evidence suggest that TE biopsy can accurately represent blastocyst karyotype after cIVF insemination.
This study had some limitations. First, all WGA protocols have limitations, such as amplification bias and error, and we performed sperm WGA using limited samples. We cannot rule out the possibility that in some specific situations, such as DNA damage, sperm can be amplified by Picoplex and ChromInst. Second, we retrospectively collected data on pregnancy outcomes after frozen PGT; therefore, the differences in fertilization and embryo development between cIVF and ICSI could not be determined. Further research is required to determine whether the patient population of PGT-A will benefit from cIVF in terms of clinical outcomes. Third, the inclusion of only 12 aneuploid blastocysts provided insufficient samples to validate the PGT-A results in the cIVF cycles.
In conclusion, we conducted a systematic study to evaluate the feasibility of using cIVF for PGT-A. Our results showed that under the WGA protocols of Picoplex and ChromInst, sperm failed to amplify, and the risk of parental contamination in the cIVF-PGT-A cycles was low. After biopsy in frozen cycles, the embryos derived from the cIVF and ICSI groups exhibited similar pregnancy rates. Given that cIVF incurs less handling of gametes, less laboratory labor load, and lower cost than ICSI, we suggest that cIVF should be the preferred insemination method for PGT-A cycles with non-male factor infertility.
Data availability
All data used and/or analyzed in this manuscript are available from the corresponding authors upon reasonable request.
References
Handyside AH, Pattinson JK, Penketh RJ, Delhanty JD, Winston RM, Tuddenham EG. Biopsy of human preimplantation embryos and sexing by DNA amplification. Lancet. 1989;1:347–9.
Palermo G, Joris H, Devroey P, Van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet. 1992;340:17–8.
Practice Committees of the American Society for Reproductive Medicine and the Society for Assisted Reproductive Technology. Intracytoplasmic sperm injection (ICSI) for non-male factor infertility: a committee opinion. Fertil Steril. 2012;98:1395–9.
ESHRE PGT Consortium and SIG-Embryology Biopsy Working Group, Kokkali G, Coticchio G, Bronet F, Celebi C, Cimadomo D, et al. ESHRE PGT Consortium and SIG Embryology good practice recommendations for polar body and embryo biopsy for PGT. Hum Reprod Open. 2020;2020:hoaa020.
Preimplantation Genetic Diagnosis International Society (PGDIS). Guidelines for good practice in PGD: programme requirements and laboratory quality assurance. Reprod Biomed Online. 2008;16:134–47.
Esteves SC, Roque M, Bedoschi G, Haahr T, Humaidan P. Intracytoplasmic sperm injection for male infertility and consequences for offspring. Nat Rev Urol. 2018;15:535–62.
Geng T, Cheng L, Ge C, Zhang Y. The effect of ICSI in infertility couples with non-male factor: a systematic review and meta-analysis. J Assist Reprod Genet. 2020;37:2929–45.
Abbas AM, Hussein RS, Elsenity MA, Samaha II, El Etriby KA, Abd El-Ghany MF, et al. Higher clinical pregnancy rate with in-vitro fertilization versus intracytoplasmic sperm injection in treatment of non-male factor infertility: systematic review and meta-analysis. J Gynecol Obstet Hum Reprod. 2020;49: 101706.
Dang VQ, Vuong LN, Luu TM, Pham TD, Ho TM, Ha AN, et al. Intracytoplasmic sperm injection versus conventional in-vitro fertilisation in couples with infertility in whom the male partner has normal total sperm count and motility: an open-label, randomised controlled trial. Lancet. 2021;397:1554–63.
Huang JX, Gao YQ, Chen XT, Han YQ, Song JY, Sun ZG. Impact of intracytoplasmic sperm injection in women with non-male factor infertility: a systematic review and meta-analysis. Front Reprod Health. 2022;4:1029381.
Ola B, Afnan M, Sharif K, Papaioannou S, Hammadieh N, Barratt CL. Should ICSI be the treatment of choice for all cases of in-vitro conception? Considerations of fertilization and embryo development, cost effectiveness and safety. Hum Reprod. 2001;16:2485–90.
Practice Committees of the American Society for Reproductive Medicine and the Society for Assisted Reproductive Technology. Intracytoplasmic sperm injection (ICSI) for non-male factor indications: a committee opinion. Fertil Steril. 2020;114:239–45.
Feldman B, Aizer A, Brengauz M, Dotan K, Levron J, Schiff E, et al. Pre-implantation genetic diagnosis-should we use ICSI for all? J Assist Reprod Genet. 2017;34:1179–83.
De Munck N, El Khatib I, Abdala A, El-Damen A, Bayram A, Arnanz A, et al. Intracytoplasmic sperm injection is not superior to conventional IVF in couples with non-male factor infertility and preimplantation genetic testing for aneuploidies (PGT-A). Hum Reprod. 2020;35:317–27.
Deng J, Kuyoro O, Zhao Q, Behr B, Lathi RB. Comparison of aneuploidy rates between conventional in vitro fertilization and intracytoplasmic sperm injection in in vitro fertilization-intracytoplasmic sperm injection split insemination cycles. F S Rep. 2020;1:277–81.
Palmerola KL, Vitez SF, Amrane S, Fischer CP, Forman EJ. Minimizing mosaicism: assessing the impact of fertilization method on rate of mosaicism after next-generation sequencing (NGS) preimplantation genetic testing for aneuploidy (PGT-A). J Assist Reprod Genet. 2019;36:153–7.
Patel K, Vaughan DA, Rodday AM, Penzias A, Sakkas D. Compared with conventional insemination, intracytoplasmic sperm injection provides no benefit in cases of nonmale factor infertility as evidenced by comparable euploidy rate. Fertil Steril. 2023;120:277–86.
World Health Organization, Department of Reproductive Health and Research. WHO laboratory manual for the examination and processing of human semen, 5th edn. Geneve: WHO Press; 2010.
Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L, Nagy ZP. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Hum Reprod. 2013;28:2298–307.
Xie P, Liu P, Zhang S, Cheng D, Chen D, Tan YQ, et al. Segmental aneuploidies with 1 Mb resolution in human preimplantation blastocysts. Genet Med. 2022;24:2285–95.
Gardner DK, Schoolcraft WB. In-vitro culture of human blastocysts. In: Jansen R, Mortimer D, editors. Towards reproductive certainty: fertility and genetics beyond 1999. Carnforth: Parthenon Press; 1999. p. 378–88.
Zhang S, Luo K, Cheng D, Tan Y, Lu C, He H, et al. Number of biopsied trophectoderm cells is likely to affect the implantation potential of blastocysts with poor trophectoderm quality. Fertil Steril. 2016;105:1222-7.e4.
Zhou S, Xie P, Zhang S, Hu L, Luo K, Gong F, et al. Complex mosaic blastocysts after preimplantation genetic testing: prevalence and outcomes after re-biopsy and re-vitrification. Reprod Biomed Online. 2021;43:215–22.
Dong Y, Liu D, Zou Y, Wan C, Chen C, Dong M, et al. Preimplantation genetic testing for human blastocysts with potential parental contamination using a quantitative parental contamination test (qPCT): an evidence-based study. Reprod Biomed Online. 2023;46:69–79.
Lynch C, Cater E, Charitou M, Forbes H, Griffin D, Gordon T. Intracytoplasmic sperm injection is not necessary as a preventive measure against paternal cell contamination in preimplantation genetic testing. Reprod Biomed Online. 2019;39(S1):e24–5.
Li H, Cui X, Arnheim N. Direct electrophoretic detection of the allelic state of single DNA molecules in human sperm by using the polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:4580–4.
Jiang Z, Zhang X, Deka R, Jin L. Genome amplification of single sperm using multiple displacement amplification. Nucleic Acids Res. 2005;33: e91.
Tran QT, Jatsenko T, Poolamets O, Tšuiko O, Lubenets D, Reimand T, et al. Chromosomal scan of single sperm cells by combining fluorescence-activated cell sorting and next-generation sequencing. J Assist Reprod Genet. 2019;36:91–7.
Berntsen S, Laivuori H, la Cour Freiesleben N, Loft A, Söderström-Anttila V, B Oldereid N, et al. A systematic review and meta-analysis on the association between ICSI and chromosome abnormalities. Hum Reprod Update. 2021;27:801–47.
Tannus S, Son WY, Gilman A, Younes G, Shavit T, Dahan MH. The role of intracytoplasmic sperm injection in non-male factor infertility in advanced maternal age. Hum Reprod. 2017;32:119–24.
Liu H, Zhao H, Yu G, Li M, Ma S, Zhang H, et al. Conventional in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI): which is preferred for advanced age patients with five or fewer oocytes retrieved? Arch Gynecol Obstet. 2018;297:1301–6.
Sunderam S, Boulet SL, Kawwass JF, Kissin DM. Comparing fertilization rates from intracytoplasmic sperm injection to conventional in vitro fertilization among women of advanced age with non-male factor infertility: a meta-analysis. Fertil Steril. 2020;113:354-63.e1.
Haas J, Miller TE, Nahum R, Aizer A, Kirshenbaum M, Zilberberg E, et al. The role of ICSI vs. conventional IVF for patients with advanced maternal age-a randomized controlled trial. J Assist Reprod Genet. 2021;38:95–100.
Li G, Yu Y, Fan Y, Li C, Xu X, Duan J, et al. Genome-wide abnormal DNA methylome of the human blastocyst in assisted reproductive technology. J Genet Genomics. 2017;44:475–81.
Chen Y, Gao Y, Jia J, Chang L, Liu P, Qiao J, et al. DNA methylome reveals the cellular origin of cell-free DNA in the spent medium of human preimplantation embryos. J Clin Invest. 2021;131: e146051.
Victor AR, Griffin DK, Brake AJ, Tyndall JC, Murphy AE, Lepkowsky LT, et al. Assessment of aneuploidy concordance between clinical trophectoderm biopsy and blastocyst. Hum Reprod. 2019;34:181–92.
Navratil R, Horak J, Hornak M, Kubicek D, Balcova M, Tauwinklova G, et al. Concordance of various chromosomal errors among different parts of the embryo and the value of re-biopsy in embryos with segmental aneuploidies. Mol Hum Reprod. 2020;26:269–76.
Acknowledgements
We thank all the patients who participated in this study. We are grateful to Lu Tan and other staff members of the PGT and IVF groups at CITIC-Xiangya for their assistance.
Funding
This study was supported by the Hunan Provincial Grant for Innovative Province Construction (2019SK4012).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
YY, AW, DZ, and SL were employed by Yikon Genomics Company, Ltd. The authors declare no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, S., Xie, P., Lan, F. et al. Conventional IVF is feasible in preimplantation genetic testing for aneuploidy. J Assist Reprod Genet 40, 2333–2342 (2023). https://doi.org/10.1007/s10815-023-02916-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02916-7