Abstract
Purpose
The purpose of this study was to evaluate the capacity of bovine granulosa cells to generate reactive oxygen intermediates in response to lipopolysaccharide. We hypothesized that granulosa cells increase reactive oxygen intermediates in response to Gram-negative lipopolysaccharide in a similar manner to immune cells.
Methods
Bovine peripheral blood mononuclear cells and granulosa cells were cultured in the presence of lipopolysaccharide. Oxidative stress was evaluated using the fluorescent marker dye CellROX, and oxidative stress-related genes were measured using real-time RT-PCR.
Results
As expected, peripheral blood mononuclear cells increased oxidative stress in response to lipopolysaccharide as measured by accumulation of the fluorescent marker dye CellROX. While granulosa cells demonstrate the capacity to increase accumulation of CellROX dye in response to a positive control menadione, lipopolysaccharide had no effect on accumulation of CellROX dye. The expression of GSR, SOD1, and SOD2 were variable in peripheral blood mononuclear cells treated with lipopolysaccharide but were consistently upregulated when co-incubated with the antioxidant, N-acetyl cysteine. The expression of oxidative stressrelated genes was not altered in granulosa cells, with the exception of elevated SOD1 following lipopolysaccharide exposure in the absence of antioxidant.
Conclusions
Combined, these data suggest that while reactive stress is important in pathogen killing and inflammation in immune cells, granulosa cells do not increase oxidative stress in response to lipopolysaccharide.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
The microenvironment of the ovarian follicle is of paramount importance to the successful development of the oocyte. During uterine infection of dairy cows, the ovary has been suggested as a peripheral target of bacterial components, altering follicle growth, estradiol synthesis, and ultimately fertility [1,2,3]. We have demonstrated that isolated bovine or human granulosa cells increase synthesis of inflammatory cytokines (IL-6) and chemokines (IL-8) in response to purified bacterial components, including lipopolysaccharide (LPS) [4,5,6]. This inflammatory response alters the developmental microenvironment of the ovarian follicle, potentially impacting oocyte quality. In fact, cows that suffer uterine infection concentrate LPS in follicular fluid and have reduced fertility even after the resolution of infection [7, 8]. In vitro studies have demonstrated that LPS exposure of ovarian tissue reduces the follicle reserve, and oocyte developmental competence is reduced due to diminished meiotic competence [4, 9, 10]. It has been demonstrated that there exist resident bacteria within follicular fluid of women which have been associated with infertility [11, 12]. Granulosa cell response to LPS is Toll-like receptor (TLR) 4 dependent and results in the activation of downstream mediators including p38 and ERK. However, bovine granulosa cells do not utilize the universal regulator of TLR inflammation, NF-κB, to induce these changes as professional immune cells do [4, 13].
Peripheral blood mononuclear cells (PMBCs) readily respond to bacterial components to increase production of inflammatory mediators via TLR activation and NF-κB signaling (reviewed in [14]). A consequence of TLR and NF-κB activation in PBMCs is the increase in reactive oxygen intermediates (ROIs). Increased production of ROI serves PBMCs to kill phagocytosed bacteria by oxidative-induced damage, in addition to regulating NF-κB-regulated cytokine production. Although there is no evidence that granulosa cells act as phagocytic cells in response to bacteria, TLR-mediated increases in inflammatory mediators may require ROI generation. Interestingly, the presence of elevated ROIs during in vitro fertilization or embryo culture reduces reproductive outcomes during artificial reproduction [15,16,17]. Many studies have demonstrated that elevated ROI in semen is associated with poorer IVF outcome [18, 19].
We hypothesized that granulosa cells increase reactive oxygen in response to Gram-negative LPS in a similar manner to immune cells. Using purified bovine granulosa cell and PBMC cultures, we aim to measure LPS-induced oxidative stress by using the fluorogenic probe CellROX and quantifying expression of oxidative stress response genes GSR, SOD1, and SOD2.
The studies described here aim to expand our understanding of how granulosa cells respond to LPS and potentially alter the developmental environment of the oocyte and subsequently impact fertility of females with uterine infection.
Materials and methods
All reagents were acquired from Fisher Scientific (Waltham, MA) unless otherwise stated.
Granulosa cell isolation and culture
Bovine ovaries were collected at a local slaughterhouse and transported directly to the laboratory in antibiotic supplemented saline. Antral follicles (4–8 mm) were aspirated using a sterile needle and syringe and collected into handling medium (Medium 199 with 0.5% bovine serum albumin (BSA) fraction V, 2 mM sodium pyruvate, 1% penicillin-streptomycin, 20 mM HEPES, and 50μg/ml heparin). Granulosa cells were isolated from 4 to 8 mm follicles to generate a homogenous population of cells that are FSH responsive and LH non-responsive [20]. Our previous studies have shown that granulosa cells from 4 to 8 mm follicles are more responsive to LPS than granulosa cells isolated from dominant follicles [4, 6]. Crude granulosa cell preparations were centrifuged at 400×g for 10 min. Subsequent cell pellets were resuspended in 2 ml of warm PBS and overlaid on a 1:1 Percoll/PBS solution and centrifuged for 30 min at 720×g to isolate granulosa cells by density centrifugation. Granulosa cells formed a distinct band following centrifugation which was collected and washed twice by centrifugation as above. Purified granulosa cells were resuspended in complete granulosa cell culture medium (Medium 199, 1% l-glutamine, 1× insulin-transferrin-selenium, 1% penicillin-streptomycin) at a concentration of 1.5 × 106 cells/ml. Granulosa cells were plated in 500 μl aliquots into 24-well culture plates and placed in humidified air incubators at 5% CO2 for 24 h prior to treatment.
Peripheral blood mononuclear cell isolation and culture
Holstein cows were housed at the University of Florida dairy research unit. All procedures were approved by the University of Florida Institutional Animal Care and Use Committee. Whole blood was collected from Holstein cows via the coccygeal vein into heparin vacutainers and centrifuged at 400×g for 10 min. The buffy coat was aspirated and washed again in PBS by centrifugation. Cell pellets were resuspended and underwent red cell lysis by addition of water before immediate isotonic balancing using 10× PBS. Cells were washed again by centrifugation and finally resuspended at 1.5 × 106 cells/ml and were plated in 500 μl aliquots into 24-well culture plates and placed in humidified air incubators at 5% CO2 for 24 h prior to treatment.
Measuring oxidative stress response to LPS by CellROX
Following equilibration, both PBMCs and granulosa cells were exposed to 10-fold increasing doses of ultrapure LPS (1 ng/ml–10 μg/ml; Invivogen, San Diego, CA) in fresh culture medium. A positive control (12.5 μM menadione) and antioxidant control (25 μM N-acetylcysteine (NAC)) were also added to cells, while NAC was added in combination with LPS. Cultures were finalized following 1 or 24 h of treatment. To measure oxidative stress, the cell permeant indicator dye CellROX was used in combination with flow cytometry. While in a reduced state, CellROX remains non-fluorescent and when oxidized, CellROX binds DNA and becomes fluorescent. Thirty minutes prior to termination of the experimental period, 3 μM of CellROX was added to each well. Adherent cells were treated with HyQTase to facilitate transfer of cells into FACS tubes for analysis. Mean fluorescence intensity (MFI) of CellROX was measured using an Accuri C6 cytometer from four independent experiments including all presented treatments. Only viable cells were evaluated following propidium iodide exclusion. In addition, cells were visualized on a Zeiss Axioplan-2 epifluorescence microscope after incubation with Hoechst 3342 to label DNA. Images in the green and blue channels were captured at the same exposure between samples.
Measuring expression of oxidative stress-related genes by real-time RT-PCR
At the completion of the treatment period, total RNA was isolated using the TRIzol method. Reverse transcription of 1 μg of RNA was performed using the Verso cDNA Synthesis Kit according to the manufacturer’s instructions. Sequencespecific primers were designed to span exon boundaries (Table 1), proven to have linear amplification efficiency of > 90% and showed product size consistent with the predicted sequence. Realtime RT-PCR was performed with a Bio-Rad CFX Connect using SYBR Green chemistry (iTaq Universal Master Mix; Bio-Rad) from four independent experiments including all presented treatments. Expression of 18S rRNA was compared between treatment groups and shown not be different, and as such was used as a suitable housekeeper gene. Data were normalized independently to 18S rRNA expression using the ΔΔCt method.
Statistical analysis
SPSS version 20.0 software was used for statistical analysis. Gene expression measured by real-time RT-PCR, and MFI were analyzed using general linear model univariate analysis, and comparisons were made with control using the Dunnett’s pairwise post hoc test and Bonferroni post hoc test between all treatments. Data was tested for normality by the Shapiro-Wilk test for normality. If data was not normally distributed, it was log-transformed for analysis and normality was confirmed by repeating the Shapiro-Wilk test for normality and observing Q-Q plots (see figure legends for transformed data sets). Data are presented as means ± SEM, and a p value of ≤ 0.05 was assumed to be statistically significant.
Results
Purity of granulosa cell cultures was assessed to quantify contaminating immune cells to ensure that measured oxidative stress was due to the target cells of interest. Granulosa cell culture contamination was assessed by measuring the expression of the immune cell marker PTPRC (formerly CD45) by RT-PCR. CYP19A1 expression was evaluated as a positive marker for granulosa cell enrichment (primers in Table 1). Granulosa cell preparations were shown to be immune cell free due to an absence of PTPRC PCR product in multiple granulosa cell preparations (Fig. 1).
Granulosa cell preparations are free of contaminating immune cells. Cultured peripheral blood mononuclear cells (PBMCs) and granulosa cells (GCs) were analyzed for the expression of the immune cell marker PTPRC and the granulosa cell marker CYP19A1 using RT-PCR. Three individual granulosa cell preparations are shown listed as GC1, GC2, and GC3
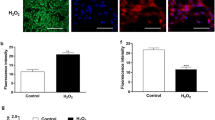
Visual fluorescence of CellROX was assessed in PBMCs and granulosa cell cultures exposed to LPS. Treatment of PBMCs with LPS for 1 h dramatically increased visual fluorescence of CellROX compared to vehicle-treated PBMCs (Fig. 2a, b). Interestingly, granulosa cells did not display any increase in CellROX fluorescence after 1 h exposure to LPS compared to vehicle-treated controls (Fig. 2c, d). As a result of this unexpected finding, we quantified LPS induced oxidative stress by measuring the MFI of CellROX in cells treated for 1 or 24 h using flow cytometry (Fig. 3). Not surprisingly, PBMCs increased CellROX MFI in response to 1 h exposure of 25 μM menadione compared to vehicletreated controls (Fig. 3a; p < 0.05). Exposure to LPS for 1 h increased CellROX MFI in PBMCs compared to vehicletreated controls, and interestingly, concurrent treatment of cells with LPS and NAC significantly reduced CellROX MFI (Fig. 3a; p < 0.05). These oxidative stress responses in PBMCs were not witnessed following treatments for 24 h (Fig. 3b). Granulosa cells increased CellROX MFI in response to 1 h exposure of 25 μM menadione when compared to vehicle-treated controls (Fig. 3c; p < 0.05). However, LPS did not increase granulosa cell CellROX MFI response after either 1 or 24 h treatment when compared to vehicle-treated controls (Fig. 3c, d).
Visualization of LPS induced oxidative stress in peripheral blood mononuclear cells and granulosa cells. Peripheral blood mononuclear cells (PBMCs; a, b) and granulosa cells (c, d) were treated with vehicle or 1 μg/ml of LPS for 1 h. Cells were visualized by epifluorescence microscopy. CellROX (green) as an indicator of oxidative stress and DNA (blue) are shown. Scale bar represents 50 μm
Quantification of oxidative stress in peripheral blood mononuclear cells and granulosa cells. Peripheral blood mononuclear cells (PBMCs; a, b) and granulosa cells (c, d) were exposed to specific treatments for a total of 1 (a, c) or 24 h (b, d). Cells were exposed to 10-fold increasing doses of ultrapure LPS alone or in combination with 25 μM of the antioxidant NAC. The oxidative agent menadione was used as a positive control. In the final 30 min of treatment, cells were exposed to 3 μM of CellROX to quantify cellular oxidative stress. Cells were analyzed by flow cytometry to quantify the mean fluorescence intensity (MFI) of 10,000 cells from each treatment. All data were log-transformed for normal distribution. Data are presented as mean ± SEM from four independent experiments. **P ≤ 0.05 compared with vehicle treated controls; ^P ≤ 0.05 compared LPS alone treated sample and LPS + NAC; analysis by general linear model univariate analysis followed by Bonferroni post hoc test
The relative expression of oxidative stress response genes (GSR, SOD1, and SOD2) were evaluated in both PBMC and granulosa cells exposed to LPS. Interestingly, following 1 h (Fig. 4) or 24 h (Fig. 5) of treatment relative expression of GSR and SOD1 was lower in PBMCs compared to granulosa cells. In PBMCs treated with menadione for 1 h (Fig. 4a–c), no change in GSR, SOD1, or SOD2 was observed. However, granulosa cells increased expression of GSR and SOD2 in response to menadione (Fig. 4d–f; not significant). Treatment of either PBMCs or granulosa cells with LPS for 1 h did not increase expression of oxidative stress response genes. There was a significant increase in expression of GSR, SOD1, and SOD2 in PBMCs co-treated with LPS and the antioxidant NAC for 1 h compared to LPS treatment alone. Conversely, there was a significant difference in SOD1 expression of granulosa cells treated with LPS in combination with NAC compared to LPS alone for 1 h. In PBMCs treated for 24 h, expression of oxidative stress response genes was highly variable (Fig. 5a–c). Expression of oxidative stress response genes in granulosa cells treated with LPS for 24 h showed no differences compared to vehicle-treated controls (Fig. 5d–f). However, expression of GSR and SOD2 in granulosa cells treated with NAC for 24 h was significantly lower compared to vehicletreated control cells.
Oxidative stress response genes following 1h treatment. Peripheral blood mononuclear cells (PBMCs; a–c) and granulosa cells (d, e) were exposed to treatments for a total of 1 h. Cells were exposed to 0.01 or 1 μg/ml of ultrapure LPS alone or in combination with 25 μM of the antioxidant NAC. The oxidative agent menadione was used as a positive control. Expression of oxidative stress gene GSR (a, d), SOD1 (b, e), and SOD2 (c, f) were measured by real-time RT-PCR. Granulosa cells GSR and SOD2 data were log-transformed for normal distribution. Data are presented as mean ± SEM from four independent experiments. **P ≤ 0.05; analysis by general linear model univariate analysis followed by Dunnett’s pairwise post hoc test or Bonferroni post hoc test
Oxidative stress response genes following 24 h treatment. Peripheral blood mononuclear cells (PBMCs; a–c) and granulosa cells (d–e) were exposed to treatments for a total of 24 h. Cells were exposed to 0.01 or 1 μg/ml of ultrapure LPS alone or in combination with 25 μM of the antioxidant NAC. The 12.5 μM of the oxidative agent menadione was used as a positive control. Expression of oxidative stress gene GSR (a, d), SOD1 (b, e), and SOD2 (c, f) were measured by real-time RT-PCR. Granulosa cells GSR and SOD2 and PBMC GSR, SOD1, and SOD2 data were log-transformed for normal distribution. Data are presented as mean ± SEM from four independent experiments. **P ≤ 0.05; analysis by general linear model univariate analysis followed by Dunnett’s pairwise post hoc test or Bonferroni post hoc test
Discussion
Granulosa cells have the capacity to elicit an innate immune response to bacterial components [4]. In a similar fashion to professional immune cells, granulosa cells increase the expression of inflammatory mediators (IL-1β, IL-6, IL-8, and TNFα) in response to bacterial cell wall components like LPS in a TLRdependent manner [4, 6, 21]. It has been demonstrated that as part of the innate immune response to pathogens, professional immune cells increase ROIs to kill pathogens and regulate production of inflammatory cytokines [22]. Here, we aimed to determine if a similar capacity to increase reactive oxygen in response to bacterial components exists in granulosa cells. We evaluated oxidative stress in PBMCs and granulosa cells following exposure to LPS, measured by accumulation of the fluorogenic probe CellROX. Our data suggest that while granulosa cells have the capacity to increase oxidative stress in response to stimulus, LPS does not increase oxidative stress or alter expression of oxidative stress response genes, GSR, SOD1, or SOD2. We had hypothesized that increased ROI in response to LPS may be a mechanisms by which oocyte quality is reduced and infertility occurs in cows suffering uterine infection.
Upon phagocytosis, professional immune cells such as macrophages increase reactive oxygen synthesis (reviewed in [23]). The production of pathogen defense ROIs begins with activation of NADPH oxidase and generation of superoxide anions which are converted to hydrogen peroxide by SOD. Increases of ROI aid in cellular killing of bacterial pathogens by oxidation within phagosomes. Interestingly, the regulation of cytokine expression (IL-1, IL-6, and TNFα) and inflammasome assembly is also effected by ROIs [13]. This observation has been linked to the participation of the key regulator of inflammatory signaling, NF-κB, which directly regulates NADPH oxidase [24]. However, we have previously shown that granulosa cells do not utilize the NF-κB pathway in response to TLR4 activation following LPS stimulation and as such may not require increased ROI to regulate cytokine production [4]. While we did not observe increases of oxidative stress in granulosa cells, this may have been due to increases in reactive oxygen response genes that have the capacity to quench free radicals, such as those involved in the glutathione synthesis pathway. However, we did not see any LPSinduced increase in GSR expression in granulosa cells which makes it unlikely that the cell is quenching any increase in ROI and more likely that cells have simply not increased ROI production in response to LPS. The reactive oxygen response genes SOD1 and SOD2 are indeed downstream targets of activated NF-κB signaling [25, 26] and involved in ROI synthesis. It may be that because granulosa cells do not utilize NF-κB signaling in response to LPS activation, which changes in SOD1 or SOD2 expression are unlikely to be seen. It is worthy of note that relative basal expression of all reactive oxygen response genes was up to 500-fold higher in granulosa cells that in PBMCs at both 1 and 24 h. This suggests that resting granulosa cells have an increased capacity to deal with elevated ROI and may not require significant gene upregulation.
It is important to note that ROIs are generated in the mitochondria as a result of normal cellular metabolism. It could be surmised that granulosa cells have a higher order of metabolic rate and oxygen consumption than PBMCs due to their non-terminally differentiated state and high proliferation capacity, while PBMCs are generally terminally differentiated. This may be evident by the numerically higher levels of oxidative stress in vehicle treated granulosa cells compared to vehicletreated PBMCs quantified by flow cytometry; indeed, the MFI of menadionetreated PBMCs is comparable to vehicletreated granulosa cells. However, this requires further investigation and the direct comparison of metabolic rates between PBMCs and granulosa cells. In addition, the physiological consequences of increased intrafollicular ROI would be detrimental to oocyte development. Indeed, during oocyte maturation when metabolic activity is highest, ROI are low [27]. It may be inferred that granulosa cells specifically do not increase oxidative stress to protect the integrity of the oocyte, protecting the germ line from oxidative damage. An alternative hypothesis is that granulosa cells require a longer period of LPS stimulation to induce acute ROI production. Indeed, granulosa cells may increase ROI in response to LPS if provided adequate time. However, the observation that granulosa cells possess the capacity to induce a rapid response to medadione stimulation is suggestive that a distinct pathway for LPSinduced ROI production exists—a paradigm which would require a complete time course exposure.
In summary, these data suggest that granulosa cells do not increase oxidative stress in the same manner as professional immune cells in response to LPS. This is surprising considering the capacity of these cells to increase expression of inflammatory mediators in response to LPS in a TLR4dependent manner [4, 5].
Conclusion
The data presented here expands on the model of how granulosa cells induce an inflammatory response to bacterial components. We conclude that while granulosa cells can produce TLR4dependent inflammation in response to LPS, they do so in the absence of increased oxidative stress. An absence of increased oxidative stress within the ovarian follicle may be a mechanism by which granulosa cells attempt to protect the developing oocyte from oxidative damage during infection.
References
Sheldon IM, Noakes DE, Rycroft AN, Pfeiffer DU, Dobson H. Influence of uterine bacterial contamination after parturition on ovarian dominant follicle selection and follicle growth and function in cattle. Reproduction. 2002;123(6):837–45.
Williams EJ, Fischer DP, Noakes DE, England GC, Rycroft A, Dobson H, et al. The relationship between uterine pathogen growth density and ovarian function in the postpartum dairy cow. Theriogenology. 2007;68(4):549–59.
Williams EJ, Sibley K, Miller AN, Lane EA, Fishwick J, Nash DM, et al. The effect of Escherichia coli lipopolysaccharide and tumour necrosis factor alpha on ovarian function. Am J Reprod Immunol. 2008;60(5):462–73.
Bromfield JJ, Sheldon IM. Lipopolysaccharide initiates inflammation in bovine granulosa cells via the TLR4 pathway and perturbs oocyte meiotic progression in vitro. Endocrinology. 2011;152(12):5029–40. https://doi.org/10.1210/en.2011-1124.
Ibrahim LA, Kramer JM, Williams RS, Bromfield JJ. Human granulosa-luteal cells initiate an innate immune response to pathogen-associated molecules. Reproduction. 2016;152(4):261–70. https://doi.org/10.1530/REP-15-0573.
Price JC, Bromfield JJ, Sheldon IM. Pathogen-associated molecular patterns initiate inflammation and perturb the endocrine function of bovine granulosa cells from ovarian dominant follicles via TLR2 and TLR4 pathways. Endocrinology. 2013;154(9):3377–86. https://doi.org/10.1210/en.2013-1102.
Bromfield JJ, Santos JEP, Block J, Williams RS, Sheldon IM. Uterine infection: linking infection and innate immunity with infertility in the high-producing dairy cow. Journal of Animal Sciences. 2015;93(5):2021–33.
Herath S, Williams EJ, Lilly ST, Gilbert RO, Dobson H, Bryant CE, et al. Ovarian follicular cells have innate immune capabilities that modulate their endocrine function. Reproduction. 2007;134(5):683–93.
Bromfield JJ, Sheldon IM. Lipopolysaccharide reduces the primordial follicle pool in the bovine ovarian cortex ex vivo and in the murine ovary in vivo. Biol Reprod. 2013;88(4):98. https://doi.org/10.1095/biolreprod.112.106914.
Sheldon IM, Price JC, Turner ML, Bromfield JJ, Cronin J. Uterine infection and immunity in cattle. In: Juengel JL, Miyamoto A, Price C, Reynolds LP, Smith MF, Webb R, editors. Reproduction in domestic ruminants VIII. Leciestershire: Context Products; 2014. p. 415–30.
Pelzer ES, Allan JA, Theodoropoulos C, Ross T, Beagley KW, Knox CL. Hormone-dependent bacterial growth, persistence and biofilm formation—a pilot study investigating human follicular fluid collected during IVF cycles. PLoS One. 2012;7(12):e49965. https://doi.org/10.1371/journal.pone.0049965.
Pelzer ES, Allan JA, Cunningham K, Mengersen K, Allan JM, Launchbury T, et al. Microbial colonization of follicular fluid: alterations in cytokine expression and adverse assisted reproduction technology outcomes. Hum Reprod. 2011;26(7):1799–812. https://doi.org/10.1093/humrep/der108.
Asehnoune K, Strassheim D, Mitra S, Kim JY, Abraham E. Involvement of reactive oxygen species in Toll-like receptor 4-dependent activation of NF-kappa B. J Immunol. 2004;172(4):2522–9.
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801.
Goto Y, Noda Y, Mori T, Nakano M. Increased generation of reactive oxygen species in embryos cultured in vitro. Free Radic Biol Med. 1993;15(1):69–75.
Guerin P, El Mouatassim S, Menezo Y. Oxidative stress and protection against reactive oxygen species in the pre-implantation embryo and its surroundings. Hum Reprod Update. 2001;7(2):175–89.
Bain NT, Madan P, Betts DH. The early embryo response to intracellular reactive oxygen species is developmentally regulated. Reprod Fertil Dev. 2011;23(4):561–75. https://doi.org/10.1071/RD10148.
Agarwal A, Allamaneni SS, Nallella KP, George AT, Mascha E. Correlation of reactive oxygen species levels with the fertilization rate after in vitro fertilization: a qualified meta-analysis. Fertil Steril. 2005;84(1):228–31. https://doi.org/10.1016/j.fertnstert.2004.12.057.
Aitken RJ, Baker MA, Sawyer D. Oxidative stress in the male germ line and its role in the aetiology of male infertility and genetic disease. Reprod BioMed Online. 2003;7(1):65–70.
Xu Z, Garverick HA, Smith GW, Smith MF, Hamilton SA, Youngquist RS. Expression of follicle-stimulating hormone and luteinizing hormone receptor messenger ribonucleic acids in bovine follicles during the first follicular wave. Biol Reprod. 1995;53(4):951–7.
Price JC, Sheldon IM. Granulosa cells from emerged antral follicles of the bovine ovary initiate inflammation in response to bacterial pathogen-associated molecular patterns via Toll-like receptor pathways. Biol Reprod. 2013;89(5):119. https://doi.org/10.1095/biolreprod.113.110965.
Ding AH, Nathan CF, Stuehr DJ. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J Immunol. 1988;141(7):2407–12.
Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev. 2015;264(1):182–203. https://doi.org/10.1111/imr.12266.
Anrather J, Racchumi G, Iadecola C. NF-kappaB regulates phagocytic NADPH oxidase by inducing the expression of gp91phox. J Biol Chem. 2006;281(9):5657–67. https://doi.org/10.1074/jbc.M506172200.
Rojo AI, Salinas M, Martin D, Perona R, Cuadrado A. Regulation of Cu/Zn-superoxide dismutase expression via the phosphatidylinositol 3 kinase/Akt pathway and nuclear factor-kappaB. J Neurosci. 2004;24(33):7324–34. https://doi.org/10.1523/JNEUROSCI.2111-04.2004.
Jones PL, Ping D, Boss JM. Tumor necrosis factor alpha and interleukin-1beta regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-beta and NF-kappaB. Mol Cell Biol. 1997;17(12):6970–81.
Morado SA, Cetica PD, Beconi MT, Dalvit GC. Reactive oxygen species in bovine oocyte maturation in vitro. Reprod Fertil Dev. 2009;21(4):608–14. https://doi.org/10.1071/RD08198.
Acknowledgements
The authors wish to thank Dr. John Driver for his support and valuable guidance in completing the flow cytometry analysis. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number R01HD084316.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Bromfield, J.J., Iacovides, S.M. Evaluating lipopolysaccharide-induced oxidative stress in bovine granulosa cells. J Assist Reprod Genet 34, 1619–1626 (2017). https://doi.org/10.1007/s10815-017-1031-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-017-1031-2