Abstract
Purpose
To establish the phenotype of reproductive aging in our mouse model. To test the hypotheses that reproductive aging is associated with a decrease in mitochondrial abundance that could ultimately reflect dysfunction in oocytes.
Methods
Breeding studies were performed in young and aged female virgin wild type C57BL6J mice to establish their reproductive phenotype by measuring time to conception, litter size, and live birth per dam. Individual oocytes were analyzed for mtDNA content. Transmission electron microscopy was used to study ultrastructure of mitochondria in oocytes.
Results
Old females were found to have significantly prolonged time to conception and fewer surviving pups in their litters. Oocytes from old mice had 2.7-fold less mtDNA compared to younger controls (p < 0.001; 95 % CI 2.1–3.5). Decrease in mitochondrial organelle abundance in old animal’s oocytes was confirmed with transmission electron microscopy. Distinct morphological changes were noted in mitochondria, suggesting altered mitochondrial biogenesis in the old animals’ oocytes.
Conclusions
Reproductive aging in mice is associated with reduced reproductive competence. Aging is associated with a significant decrease in number of mitochondria in oocytes. Our data support mitochondrial organelle loss and dysfunction in oocytes as a potential etiology for reproductive senescence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rapid decline of fertility in aging women is accompanied by an increase in miscarriage rates and increased risk of aneuploidy in offspring, yet the mechanisms are poorly understood. The main mechanism involves age-related accumulation of chromosomal defects in oocytes [13]. Oocyte aneuploidy results primarily from premature separation of sister chromatids during meiosis I or from whole chromosome nondisjunction during meiosis II [1, 8].
Oocytes are the largest cells and have the most mitochondrial DNA (mtDNA) of any cell in the organism [11]. They require substantial energy to support transcription and translation during maturation which is generated through mitochondrial electron transport chain (ETC) activity. Mitochondria participate in calcium homeostasis, oxygen sensing, fatty acid oxidation, signal transduction [23], as well as, steroid and heme synthesis, oxidative stress and the regulation of apoptosis [26]. Deficiency of ETC proteins depletes cellular ATP and compromise spindle formation, checkpoint control and chromosome alignment, with negative affect on chromosome segregation, likelihood of embryo implantation and viability. Low mitochondrial membrane potential is associated with chromosome non-disjunction, aneuploidy and mosaicism in human embryos [23]. Studies that correlate ATP levels [24] with normal oocyte meiotic and mitotic spindle organization and chromosomal segregation confirm a critical role for mitochondrial function [18]. Mitochondrial distribution in oocytes and early embryos is closely correlated with developmental competence [27]. Therefore mitochondria play a major role in mechanisms that govern oocyte quality and quantity.
Previous research, recently reviewed by Bentov et al., has only partially defined how mitochondrial number and function in oocytes changes with maternal aging [2]. It is important to delineate whether aging leads to quantitative or qualitative changes or both in oocyte mitochondria. If female reproductive aging is demonstrated to be primarily a cytoplasmic or mitochondrial process existing techniques that involve either chromosome transfer or cytoplasmic transfer can be utilized to enable older females and those with mtDNA diseases to reproduce [21]. To address this knowledge gap, we established the phenotype of reproductive aging in wild type C57BL6J mice. This model served to test the hypothesis that reproductive aging is associated with a decrease in mitochondrial abundance in oocytes.
Materials and methods
Procedures were fully compliant with Emory IACUC and NIH guidelines.
Breeding
Wild type (WT) female C57BL6J mice of varying age were mated with young WT males with previously proven fertility. Harems included four virgin females per one young male. Mice were allowed to cohabitate for 20 days, at which point the male was removed and the females were separated into individual cages. Reproductive phenotype was operationally defined by time to conception (delivery date minus the duration of typical mouse gestation—21 days), litter size, and live birth per dam measured at birth, one and three postnatal weeks.
Oocyte collection
Seven mice per younger age group and 15 mice >300 days underwent superovulation with 4 IU of pregnant mare’s serum gonadotropin (PMSG) injected intraperitoneally (IP). Ovulation was triggered by 2.5 IU of human chorionic gonadotropin (hCG) IP 48 h after PMSG. Mice were humanely euthanized 14 to 16 h after hCG injection. The oviducts were then dissected and ovulated oocytes were isolated under a stereomicroscope in M2 media. Cumulus cells were dissociated using hyaluronidase, Metaphase II (MII) oocytes were washed 3 times in M2 media. A pipette was used to transfer individual oocytes.
Quantitation of mtDNA using real-time PCR
DNA was extracted from individual MII oocytes by three rounds of freeze-thawing (heating at 99 °C, followed by immediate freezing). mtDNA amplification was performed using real-time PCR techniques adapted from Cote et al. [4, 6]. Mouse mitochondrial cytochrome oxidase subunit 1 was the target for mtDNA. Amplification was performed in a LightCycler 480 (Roche Diagnostics, Indianapolis, IN). Standard DNA curves for quantification of the products were generated using serial dilutions of the mitochondrial target sequences. Amplification efficiency of quantitative real-time PCR was calculated to be 1.8 using standard curves.
Ultrastructural evaluations with transmission electron microscopy (TEM) analysis
Following superovulation as described above but just prior to ovulation mice were humanely euthanized and their ovaries were dissected out and fixed. Fixation of ovaries was based on a protocol applied for heart tissue fixation previously used in our lab [9]. The samples are perfusion-fixed at 50 mm Hg constant pressure (monitored) using 10 % neutral buffered formalin or diluted Karnovsky’s fixative. The perfusion-fixed sample was cut into 2 mm slices. Samples were sectioned longitudinally and transversely, and fixed in 2 % glutaraldehyde-cacodylate for 2 h at 4 °C. Tissue is rinsed in cold Ringer’s solution and post-fixed in 1 % OsO4 (Sigma, St. Louis, MO) in PBS, ph 7.4 for 2 to 3 h. Following osmication and rinses, tissue was dehydrated with graded ethanols, and embedded in resin [20]. Samples were sectioned (100 nm), stained with uranyl acetate, and examined on a JEOL-JEM-100CX electron microscope. (JEOL, Tokyo, Japan). Both ovaries from four animals per age group (age 79 days and 320 days) were analyzed. For each embedded sample, 10 to 15 sections of entire mature oocytes were selected and photographed at a uniform magnification (11,000x) by a single investigator. Photomicrographs were enlarged to 8 × 10-inch prints and reviewed independently by two investigators for mitochondrial structure [5]. Mitochondrial number per uniform high power field were also observed for the two groups.
Statistical analysis
Data were analyzed by using a t-test for continuous distributions. Multiple means were compared by one-way ANOVA followed by a Tukey’s post hoc test. The statistical significance levels for all tests were set at ≤0.05. All data are reported as means with their associated standard deviations. Relative risk and 95 % confidence intervals are shown where appropriate. All data management and statistical analyses were implemented in SAS Package Version 9.2 (SAS Institute, Inc., Cary, North Carolina).
Results
Reproductive phenotype
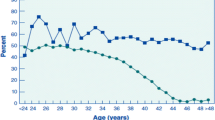
To delineate the effect of age on reproduction virgin WT young females (n = 26, mean age: 75 ± 5 days) and old females (n = 14, mean age 203 ± 22 days) were bred with young fertile WT males. Old females were found to have significantly prolonged time to conception (p < 0.001). While the litter size was similar for young and old dams, old dams had significantly fewer live born pups. Fewer pups born to old dams survived until weaning compared to pups born to young dams. Age was significantly negatively correlated with number of live born pups (p = 0.02), pups alive 1 week postnatal (p < 0.001), and those alive 3 weeks postnatal (p = 0.02) (Fig. 1).
Fertility by female age. Female fertility in C57BL6J mice decreases with age. Old WT dams took longer to conceived when mated with a young fertile WT male. While the litter size was similar for young and old dams, old dams had fewer live born pups. Fewer pups born to old dams survived until weaning compared to pups born to young dams. *p value ≤ 0.05
mtDNA in oocytes
Real time PCR was used to quantitate mtDNA in individual oocytes. mtDNA abundance was significantly decreased in oocytes from older females as compared to controls (Fig. 2). Oocytes from mice older than 300 days had 2.7-fold less mtDNA compared to younger mice (p < 0.001; 95 % CI 2.1–3.5). There was no significant difference in mtDNA abundance in any of the age groups younger than 300 days.
Morphology of mitochondria in oocytes
The morphology of MII oocytes of young and old mice was examined by TEM. TEM indicated distinct changes in mitochondrial morphology with age. This analysis indicates a decrease in mitochondrial organelle abundance per magnification field in old animals oocytes compared to young controls (p < 0.001). In young control animals oocyte mitochondria are undifferentiated, round in shape, with few cristae, and an electron dense matrix. The mitochondria from old animals have numerous cristae and appear more differentiated, while small and undifferentiated mitochondria are rare compared to young controls. While most mitochondria have a round shape in both young and old animals; elongated organelles are occasionally seen in the older animal but are completely absent in the young mice (Fig. 3). These findings suggest altered mitochondrial biogenesis in the old animals’ oocytes with more rapid differentiation, yet overall fewer organelles. Vacuoles were more commonly observed in the ooplasm mitochondria of young mice compared to old mice; while the role of these vacuoles is unknown they have been observed in prior studies [14].
TEM of mitochondria from young and old mouse oocytes. MII Oocytes from hyperstimulated young and old mice were analyzed by TEM mitochondrial structure was compared. Representative electron micrographs of ooplasm at 11,000x magnification are shown; higher magnification views of individual mitochondria are also presented. A. abundant mitochondria per field also note different size mitochondria compared to B. B. relatively few mitochondria of uniform size per field in ooplasm of an aged animal. C & E. undifferentiated round mitochondria with an electron dense matrix vs. D, F & H. more differentiated mitochondria with an elongated shape and distinct cristae G. arrow indicating vacuoles within mitochondria
Discussion
Our data establish the reproductive phenotype in aging C57BL6J mice. Fecundity is overall lower with advancing age while those that do get pregnant are less likely to deliver viable offspring. This finding is consistent with the reproductive phenotype of older human females and serves as a model for further studies of reproductive senescence. While a number of prior studies have examined mtDNA levels and mitochondrial ultrastructure in oocytes, no prior study has attempted to systematically address mtDNA abundance and mitochondrial structure as a function of reproductive aging. Our data demonstrate that aging is associated with a significant decrease in mtDNA content in oocytes. TEM confirms our mtDNA data and indicates a decrease in organelle abundance in oocytes from old mice. Finally, we describe distinct morphological changes in mitochondria of old animals’ oocytes, which suggest altered mitochondrial biogenesis.
In our study oocytes from old animals had 2.7-fold less mtDNA compared to younger mice. The oocyte mtDNA copy number threshold critical for normal postimplantation development was determined to be one third of the normal mtDNA content [25]. Our finding is consistent with existence of a critical postimplantation developmental threshold for the number of copies of mtDNA in the mature oocyte. Therefore, reproductive aging is associated with the same degree of mtDNA loss in oocytes previously found to be critical for reproductive success. This finding indicates that oocytes from old animal may be at a relative disadvantage via the mitochondrial genetic bottle neck. mtDNA copy number has been previously linked to fertility potential in human oocytes. Oocytes from patients with ovarian insufficiency have been shown to have decreased mtDNA content [10]. Low mtDNA abundance in oocytes has been associated with a decreased fertilization potential [15, 16]. Oocytes from older women are more likely to harbor the 0.5-kb “common” deletion, indicating that aging is also associated with mtDNA mutagenesis [3, 7]. Cumulative mitochondrial damage with age may be attributed to oxidative stress generated by ROS from the mitochondria’s own basal metabolism [19]. Interestingly we only noted a significant decrease in mtDNA content in mice older than 300 days; however, reproduction was compromised in animals older than 200 days. Unfortunately mice in the 200–300 day age group did not stimulate well and produced fewer oocytes compared to younger mice, this led to a wide standard deviation in mtDNA abundance in this group. To compensate for poor response to stimulation additional animals were stimulated in >300 day old group to generate an adequate number of oocytes for analysis.
Our data indicates that old animals’ oocytes contain fewer mitochondria and that their organelles are more differentiated; compared to young animals they have few undifferentiated mitochondria. Prior human studies have reported that advanced maternal age is associated with decreased inner mitochondrial membrane potential, and this adversely affects fertilization and early developmental potential of oocytes [27, 28]. Since mtDNA codes for several subunits of respiratory chain complexes decreased mtDNA can lead to decreased ATP-generation. Increased oxidative stress in aged murine oocytes has been linked to low ooplasmic ATP levels [22]. Undifferentiated appearance of mitochondria in MII oocytes has previously been described; therefore, our finding is consistent with prior reports [12, 17]. In MII oocytes mitochondria have a round shape with few well defined cristae. Differentiated mitochondria appear more akin to those found in somatic cells—the organelles become elongated, distinct cristae are abundant. The observed lack of undifferentiated mitochondria in old animals oocytes in our study is consistent with age related decrease in mitochondrial function described in prior studies. This may underlie impaired oocyte development, nuclear spindle activity, and ultimately abnormal chromosome segregation seen in older mammals.
The relationship between mitochondrial number, mtDNA abundance and function in oocytes of aging females needs further clarification with respect to their individual and combined impact on reproductive competence. Since women are increasingly delaying reproduction beyond age 35, this line of research has important implications for pathogenesis of common diseases due to inheritence of dysfunctional mitochondria by the offspring and developmental origins of health and disease. Additionally, understanding the exact role of mitochondria in reproductive senescence may help in generating potential therapeutic targets for treatment of age related infertility and pregnancy loss due chromosome aneuploidy. Future directions include quantifying effects of aging on mitochondrial activity and protein expression in oocytes.
References
Angell R. First-meiotic-division nondisjunction in human oocytes. Am J Hum Genet. 1997;61(1):23–32.
Bentov Y, et al. The contribution of mitochondrial function to reproductive aging. J Assist Reprod Genet. 2011;28(9):773–83.
Chen X, et al. Rearranged mitochondrial genomes are present in human oocytes. Am J Hum Genet. 1995;57(2):239–47.
Cote HC, et al. Mitochondrial: nuclear DNA ratios in peripheral blood cells from human immunodeficiency virus (HIV)-infected patients who received selected HIV antiretroviral drug regimens. J Infect Dis. 2003;187(12):1972–6.
Dalakas MC, et al. Mitochondrial myopathy caused by long-term zidovudine therapy. N Engl J Med. 1990;322(16):1098–105.
Davani EY, et al. Insulin-like growth factor-1 protects ischemic murine myocardium from ischemia/reperfusion associated injury. Crit Care. 2003;7(6):R176–83.
Keefe DL, et al. Mitochondrial deoxyribonucleic acid deletions in oocytes and reproductive aging in women. Fertil Steril. 1995;64(3):577–83.
Kuliev A, et al. Chromosomal abnormalities in a series of 6,733 human oocytes in preimplantation diagnosis for age-related aneuploidies. Reprod Biomed Online. 2003;6(1):54–9.
Lewis W, et al. Mitochondrial ultrastructural and molecular changes induced by zidovudine in rat hearts. Lab Invest. 1991;65(2):228–36.
May-Panloup P, et al. Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum Reprod. 2005;20(3):593–7.
Michaels GS, Hauswirth WW, Laipis PJ. Mitochondrial DNA copy number in bovine oocytes and somatic cells. Dev Biol. 1982;94(1):246–51.
Motta PM, et al. Mitochondrial morphology in human fetal and adult female germ cells. Hum Reprod. 2000;15 Suppl 2:129–47.
Pellestor F, et al. Maternal aging and chromosomal abnormalities: new data drawn from in vitro unfertilized human oocytes. Hum Genet. 2003;112(2):195–203.
Perez GI, et al. Genetic variance modifies apoptosis susceptibility in mature oocytes via alterations in DNA repair capacity and mitochondrial ultrastructure. Cell Death Differ. 2007;14(3):524–33.
Reynier P, et al. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod. 2001;7(5):425–9.
Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril. 2006;85(3):584–91.
Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15 Suppl 2:148–59.
Schon EA, et al. Chromosomal non-disjunction in human oocytes: is there a mitochondrial connection? Hum Reprod. 2000;15 Suppl 2:160–72.
Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A. 1994;91(23):10771–8.
Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26(1):31–43.
Tachibana M, Sparman M, Mitalipov S. Chromosome transfer in mature oocytes. Nat Protoc. 2010;5(6):1138–47.
Tarin JJ. Potential effects of age-associated oxidative stress on mammalian oocytes/embryos. Mol Hum Reprod. 1996;2(10):717–24.
Van Blerkom J. Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction. 2004;128(3):269–80.
Van Blerkom J, Davis PW, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod. 1995;10(2):415–24.
Wai T, et al. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod. 2010;83(1):52–62.
Wang W, et al. Superoxide flashes in single mitochondria. Cell. 2008;134(2):279–90.
Wilding M, et al. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum Reprod. 2001;16(5):909–17.
Wilding M, et al. Chaotic mosaicism in human preimplantation embryos is correlated with a low mitochondrial membrane potential. Fertil Steril. 2003;79(2):340–6.
Acknowledgments
These data were presented at the 59th annual Society for Gynecologic Investigation meeting. Supported by DHHS, NIH DA030996 to William Lewis MD. Authors have nothing to disclose
Author information
Authors and Affiliations
Corresponding author
Additional information
Capsule
Reproductive aging in mice is associated with reduced reproductive competence, a significant decrease in number and altered structure of ooplasmic mitochondria.
Rights and permissions
About this article
Cite this article
Kushnir, V.A., Ludaway, T., Russ, R.B. et al. Reproductive aging is associated with decreased mitochondrial abundance and altered structure in murine oocytes. J Assist Reprod Genet 29, 637–642 (2012). https://doi.org/10.1007/s10815-012-9771-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-012-9771-5