Abstract
Biological soil crusts (BSCs) represent a micro-ecosystem consisting of various microorganisms including algae in drylands worldwide. The green algal genus Klebsormidium is a typical member of such communities, and because of its filamentous morphology and sticky cells, it plays an ecological key role in the stabilization of soil surfaces. In the present study, we investigated for the first time the phylogeny and ecophysiological performance of five BSC Klebsormidium strains from the semi-arid Colorado Plateau, USA. The molecular phylogeny of rbcL sequences showed that these strains belong to two subclades, which have been described before from mainly humid habitats. During controlled dehydration, the effective quantum yield of photosystem II decreased during 370–430 min of exposure. After controlled rehydration, all strains recovered between 32.9 and 97.6% of the control, but with significant differences depending on the genetic lineage. All five isolates grew between 18.1 and 27.9 °C, with isolate-specific optimal growth temperatures. Similarly, all strains were grown under increasing photon fluence rates from 4 to 110 μmol photons m−2 s−1 with optima under the higher tested light levels. Two strains showed inhibition at higher photon fluence rates and hence low light requirements for growth. Photosynthesis under increasing photon fluence rates up to 314 μmol photons m−2 s−1 indicated again low light requirements, but with no photoinhibition. All results underline a pronounced plasticity of the ecophysiological traits of the investigated Klebsormidium strains, which allow these terrestrial algae to exploit the environmental gradients within BSCs in arid regions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In drylands, where growth of vascular plants is limited due to harsh environmental conditions, biological soils crusts (BSCs) play an important ecological component of the habitat. BSCs typically consist of bacteria, cyanobacteria, non-lichenized fungi, lichens, bryophytes, algae, and microfauna (Belnap 2003). This micro-community provides important ecosystem functions, making them to real ecosystem engineers. Through the BSC composition of soil particles and organic compounds, they stabilize soils and influence water cycles. As BSCs consist of various photoautotrophic organisms, atmospheric carbon dioxide is fixed, biologically transformed, and finally transferred into the soil. In addition, atmospheric nitrogen can be fixed by cyanobacteria, which contributes to nutrient enrichments (Belnap 2002).
Drylands cover up to 40% of the terrestrial land surface (Belnap et al. 2016), and they consist of regions with hyper-arid, arid, semi-arid, and dry conditions, which includes polar deserts as well. In these regions, BSCs can cover up 70–90% of the terrestrial surface (Belnap 1994; Williams et al. 2017). Because of their biological activity, it has been estimated that these communities contribute ∼7% of the net global primary production and ∼50% of the global nitrogen fixation (Elbert et al. 2012). In fact, these numbers might be even higher, as previous studies underestimated the biological activity, due to, for example, experimental biases of chlorophyll fluorescence-based methods for photosynthesis (Raanan et al. 2016).
BSCs have their main distribution area in North America in southwestern hot and cold deserts. During the last decades, investigations focused mainly on occurrence, biodiversity, function, and succession of BSCs in the USA, rather than on ecophysiological aspects of single components. The cool semi-arid Colorado Plateau, localized in southwestern USA, is characterized by average summer temperatures of 16–26 °C and average winter temperatures between 0 and 8 °C, accompanied with temporal annual soil freezing (Rosentreter and Belnap 2003). Precipitation is about 150–250 mm per year, with 35–40% as summer monsoons and cold winters with significant snow input (Rosentreter and Belnap 2003). Vascular plants on the Colorado Plateau are dominated by Coleogyne, Atriplex, Artemisia, Pinus, and Juniperus in a mixed shrub-, grass-, and woodland (Rosentreter and Belnap 2003). BSCs on the Colorado Plateau are widely distributed between the loosely distributed vascular plants. They are composed of cyanobacteria, green algae, Bacillariophyceae, bryophytes, and lichens. The green algal diversity is high but usually low in biomass (Rosentreter and Belnap 2003). Klebsormidium flaccidum is a common green algal species in these BSCs (Rosentreter and Belnap 2003). Although Klebsormidium seems to have low biomass in these particular BSCs, it is an alga which actively supports the formation of crusts by entrapping soil particles due to their filamentous morphology and secretion of sticky mucilage (Büdel et al. 2016).
Members of the genus Klebsormidium (Klebsormidiophyceae, Streptophyta) are worldwide distributed in different habitats such as natural and artificial substrates, including rock surfaces, soil, and BSCs (e.g., Lokhorst 1996; Rindi et al. 2011; Mikhailyuk et al. 2015). The phylogeny based on ITS rRNA and rbcL sequences indicate the existence of seven well-supported superclades (A-G; Rindi et al. 2011). Superclade A contains the genus Interfilum and superclades B-G the genus Klebsormidium. The most common monophyletic clade of Klebsormidium is E, with at least 14 lineages (Rindi et al. 2011; Mikhailyuk et al. 2015; Ryšánek et al. 2015). Members of this clade can be found in many different regions, as well as in various terrestrial and freshwater habitats. Although the superclades are genetically well-supported, the morphological identification of single lineages is still challenging due to overlapping morphological characteristics and plasticity of species. However, certain genotypes can be linked to special environmental factors, e.g., acidic soils (Škaloud et al. 2014) and natural vs. artificial subaerial substrates (Škaloud and Rindi 2013). These genotypes point to the existence of cryptic species and might be suitable for a discrimination of different ecotypes.
Ecophysiological studies on Klebsormidium from BSCs focused so far mainly on strains from Europe (Glaser et al. 2017; Karsten and Rindi 2010; Karsten and Holzinger 2012) and rarely from other regions, such as Alaska (Karsten et al. 2016). Studies on strains from North American dryland BSCs are completely missing. As a recent comprehensive study of Klebsormidium strains from temperate habitats revealed an unexpectedly high physiological plasticity, strains from more extreme habitats might give new insights in the existence of infraspecific differentiation (Donner et al. 2016).
The aim of the present study was for the first time an ecophysiological characterization of five strains of Klebsormidium originating from semi-arid dryland BSCs in southwestern North America. One goal was to examine the genetic diversity of the selected strains using rbcL sequences, since such genetic data from North American BSC Klebsormidium strains are missing. Furthermore, photosynthetic activity response was evaluated as a function of a light gradient, as well under controlled desiccation and rehydration conditions. Additionally, the growth response as a function of temperature and light gradients was investigated.
Material and methods
Strain origin and culture conditions
The strains were isolated from BSCs collected in May 2015. Three sites in southwestern USA were sampled: (1) Dead Horse Point, Utah (D1; 38°28′16.01″N, 109°44′27.08″W); (2) near Moab, Utah (M1, M3; 38°38′45.57″N, 109°44′10.05″W); and (3) Grand Canyon, Arizona (G6, G7; 36°3′57.17″N, 112°8′7.30″W). All sites are located in the Colorado Plateau, a cold semi-arid dryland spanning 210,000 km2 of Utah, Colorado, Arizona, and New Mexico. The sampled sites represent typical BSCs in semi-arid drylands, where the vegetation is dominated by a patchy distribution of shrubs and grasses. Detailed data on habitat, origin, and meteorology are summarized in Table 1.
All samples were cultivated on 1.5% agar supplemented with modified Bold’s Basal Medium (3NBBM; Starr and Zeikus 1993). Detected Klebsormidium filaments were transferred to Petri dishes with fresh 3NBBM medium. After three transfers, the obtained cultures were inspected for unialgality by examination under a light microscope. Samples and unialgal stock cultures of Klebsormidium were maintained in 3NBBM at 20 °C under white fluorescent photon fluence rates of 30–50 μmol photons m−2 s−1 provided by 18 W cool tubes (Philips TLD 18 W/33, Netherlands), with a 14:10 h light/dark cycle. All experiments were carried out with vital log-phase cultures (c. 1–2 mg chlorophyll a L−1).
Light microscopy
The morphology of 4-week-old cultures was examined on 1.5% agar plates with 3NBBM medium and in liquid 3NBBM medium during the exponential growth phase. Morphology of strains was investigated using a light microscope (BX51, Olympus, Japan) with Nomarski differential interference optics. Light micrographs were taken with an Olympus UC30 camera attached to the microscope and processed with the software cellSens Entry (Olympus, Japan).
Molecular analyses
DNA was isolated according to the protocol of Ryšánek et al. (2015) and afterwards stored at −20 °C. The sequences of the rbcL gene, which encodes the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, were obtained by using polymerase chain reaction (PCR) amplification with a Touchgene Gradient cycler (Techne, United Kingdom). The rbcL gene was amplified by using the newly designed primer KF590 (5′-GAT GAA AAC GTA AAC TCT CAG C-3′) and the primer rbcL-KR2 (5′-GGT TGC CTT CGC GAG CTA-3′) (Škaloud and Rindi 2013). The primers were designed specifically to amplify Klebsormidium species. Each 20 μL reaction for PCR was conducted as described by Ryšánek et al. (2015). The PCR protocol followed that of Škaloud and Rindi (2013). Sequencing reads were assembled and edited by using SeqAssem software (Hepperle 2004).
Newly obtained Klebsormidium rbcL sequences and the sequences available in the GenBank database were used to produce an alignment for phylogenetic analyses. The final alignment of 732 base pairs was constructed by using ClustalW (Thompson et al. 1994) with MEGA v6.06 (Tamura et al. 2013). The aligned dataset was analyzed by using Bayesian analysis (BI) with MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001), maximum likelihood analysis (ML) with GARLI (Zwickl 2006), and maximum parsimony (MP) analysis with PAUP v4.0b10 (Swofford 2002). The evolutionary model was determined by using PAUP/MrModeltest v2.3 (Nylander 2004). The model selected under the Akaike information criterion was GTR + I + G. The BI analysis was performed by using the priors set as default in MrBayes; the robustness of the tree topologies was assessed by bootstrapping the dataset as described by Škaloud and Rindi (2013).
Photosynthesis under a light gradient
The effect of rising photon fluence rates on the relative electron transport rate of photosystem II (PSII) (rETR) was followed by using non-invasive pulse amplitude modulation (PAM) fluorometry. The rETR was measured with a PAM 2500 (Heinz Walz GmbH, Germany) at 22 °C. The range was chosen as previous experiments indicated that higher photon fluence rates inhibited the photosynthesis and was lethal for the cultures (data not shown). Log-phase cultures were concentrated on four replicate GF/F Whatman glass fiber filters (Whatman, Germany). The filters were kept moist with some drops of 3NBBM medium and dark-incubated for 15 min before algal cells were exposed to 14 photon flux densities (PFDs) for 2 min each ranging from 0 up to 314 μmol photons m−2 s−1. The actinic light was provided by a red LED (650 nm) of the PAM2000. The distance between fiber optic and filter surface was always kept constant at 5 mm. After each light exposure, a saturating pulse was given to detect Fm (maximum fluorescence yield) and ΔF/fm′ (effective photochemical efficiency). The relative electron transport rate of PSII (rETR) was calculated according to rETR = ΔF/fm′ * PFD where ΔF/fm′ = the effective PSII quantum efficiency. Photosynthesis-irradiance (PI) curves as rETR vs. PFD were calculated and again fitted by the mathematical photosynthesis model of Walsby (1997).
Growth experiment under a temperature and light gradient
The increase of in vivo chlorophyll a fluorescence Ft over time was used as an indicator for biomass accumulation and hence for estimation of the growth rate (Gustavs et al. 2009). For the measurement of the effect of different temperatures on the growth, cultures were grown in quadruples in 24-well microplates (Costar, Corning GmbH, Germany) with each well filled with 1.5% agar of modified 3NBBM. For the temperature gradient growth experiment, six different temperatures were applied (18.1, 19.1, 21.0, 24.6, 26.3, and 27.9 °C) at 40 μmol photons m−2 s−1 and a light/dark cycle of 16:8 h L/D.
For the temperature gradient growth experiments, a modified self-constructed algal incubator as described in Woelfel et al. (2014) was used. The adjusted incubator was equipped with a LED array (LED neutral white Ediline III 3.5 W COB Modul, Edison Opto Corporation, Taipei, Taiwan). All measurements were done with log-phase cultures, which were adapted for 10 days to the temperature conditions described above. After this pre-incubation, 200 μL of the cell suspension (c. 0.1–0.2 mg chlorophyll a L−1) was transferred onto the agar medium in a new 24-well microplate with quadruples for each strain. The chlorophyll a fluorescence as proxy for growth was measured every 24 h for 10 days with a SpectraMax M2e multiplate reader (MPR; Molecular Devices, Germany). For chlorophyll a fluorescence, an excitation wavelength of 480 nm was chosen and the emitted fluorescence was detected at a wavelength of 680 nm, using the top read function. Increasing fluorescence values were measured every 24 h for 10 days as relative fluorescence units (RFU). The fluorescence measured directly after the inoculation of the 24-well microplates served as a starting value. Before each measurement, a dark incubation of 10 min was performed in order to open all reaction centers of photosystem II. The calculation of the growth rate for each individual replicate was performed according to the well-established protocol of Gustavs et al. (2009) which requires a linear relationship between algal biomass and chlorophyll a fluorescence as well as exponential growth of the investigated algae. This method offers a simple, rapid, non-invasive, reproducible, and calibration-free measurement of growth rates in unialgal cultures, and the low detection limits avoid self-shading and nutrient limitation during growth rate determination.
The light gradient experiment was applied for six different photon fluence rates (4, 8, 15, 30, 85, and 110 μmol photons m−2 s−1) at 20 °C (±1 °C) and a light/dark cycle of 16:8 h L/D. The pre-cultivation, preparation, and measurements were the same as for the temperature gradient growth experiment (described above).
During the exponential growth phase of the Klebsormidium strains, the fluorescence Ft (RFU) at a given time point t can be calculated as Ft = F0 e μt with F0 (RFU) as initial fluorescence and μ (day−1) as growth rate in the respective time interval. For the calculation of μ, the measured chlorophyll a fluorescence values were fitted with the growth equation mentioned above. The fitting was based on the sum of the mean square error AFl which was calculated as (Ft − Ft,cal)2 with Ft (RFU) as measured fluorescence at a given time point t and Ft,cal (RFU) as calculated fluorescence at a given time point t with at least three subsequent fluorescence values. MS Excel 2013 add-in Solver with the model “GRG-non-linear” was used to minimize AFl. The growth rate μ is a relative unit and a rate of 1 equals a doubling of biomass within 1 day.
Desiccation and rehydration experiment
For the desiccation experiments, a standardized set-up was applied to follow kinetics of controlled dehydration and subsequent rehydration on the effective quantum yield of photosystem II (PSII) using non-invasive pulse amplitude modulation (PAM) fluorometry. A modified desiccation chamber after Karsten et al. (2014) was used for the experiments. As all samples originated from BSCs, all PAM measurements were done on low-light acclimated samples (30–40 μmol photons m−2 s−1). Cells of each Klebsormidium strain were concentrated on four replicate GF/F Whatman glass fiber filters (Whatman, Germany). Onto each filter, 200 μL of the cell suspension (c. 1–2 mg chlorophyll a L−1) was concentrated in the center as a light green spot. Prepared moist filters were positioned on perforated metal grids on top of a glass Petri dish placed on four glass columns inside a transparent 200-mL polystyrol box. Each box was filled with 100 g of freshly activated silica gel (Silica Gel Orange, Carl Roth, Germany) in order to create a relative air humidity of ∼10% and sealed with a transparent top lid. The boxes were kept at 22 ± 2 °C and 30–40 μmol photons m−2 s−1 (Osram Daylight Lumilux Cool White lamps L36 W/840, Osram, Germany).
The effective quantum yield (ΔF/fm′) of photosystem II was regularly determined during the dehydration period (up to 430 min depending on the strain) using a pulse-amplitude modulated fluorimeter (PAM 2500, Heinz Walz GmbH, Germany). ΔF/fm′ was calculated as (Fm′ − F)/fm′ with F as the fluorescence yield of light-treated algal cells and Fm′ as the maximum light-adapted fluorescence yield after employing an 800-ms saturation pulse as described by Schreiber and Bilger (1993). The PAM light probe was positioned outside the cover lid of the boxes (always 2 mm distance) to guarantee undisturbed RAH conditions inside, i.e., all fluorescence measurements were done through the polystyrol lids. The distance from the PAM light probe to the algal sample onto the glass fiber filters was always kept constant at 10 mm.
After the dehydration period, the dried glass fiber filters were transferred to a new polystyrol box which was filled with 100 mL tap water instead of silica gel to create a high-humidity atmosphere (>95%). The filters were rehydrated by adding 200 μL of the standard growth medium (BBM, see above) to each filter, and recovery of (Fm′ − F)/fm′ was followed with the same methodology as described above in regular intervals with the last measurement 24 h after the experimental begin.
Statistical analysis
All statistical analyses were done with the software package R (R Development Core Team 2009). Statistical significance of the median of recovery rates, growth rates under temperature and light gradient, and rETR were tested with a one-way ANOVA followed by a Tukey’s multiple comparison test to find significant differences between species, temperature, and light steps, respectively.
Results
Light microscopy
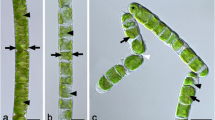
Descriptions and images of semi-arid BSC Klebsormidium strains are presented in Table 2 and Fig. 1. The multicellular algae were unbranched and consisted of filaments, either as long filament or as short fragments. Some strains with long filaments disintegrated in liquid culture into short to unicellular fragments (strains M1, G6 and G7). The length of the cells varied between 7.2 to 12.3 μm and their width between 6.2 and 8.4 μm. All cells contained plate-shaped chloroplasts, with wavy to richly waved margins. The chloroplast encircled 40–70% of the inner cell surface, with slightly lower values in liquid cultures. The pyrenoid inside the chloroplast was average to big in size, surrounded by several starch grains. Only strain D1 showed during cell division on solid medium the specific cell wall structures of H-pieces, which is an important morphological character for some Klebsormidium species (Mikhailyuk et al. 2014).
Molecular analyses
The phylogenetic analysis of Klebsormidium revealed seven superclades sensu Rindi et al. (2011) with moderate to high support (indicated with letters A–G; Fig. 2). Within superclade E sensu Rindi et al. (2011), five clades with highest support were recovered (E1, E2, E3, E4, and E5). Additionally, within superclade E, 13 lineages sensu Škaloud and Rindi (2013) were recognized (indicated as 1–13). The genetic diversity of the five newly investigated strains was low, as they are all member of the highly supported superclade E. However, the samples represent two different genotypes, inferring with two distinct lineages. The strains M1, G6, and G7 belonged with high support to lineage E2 (Škaloud and Rindi 2013). Other members of this lineage were described so far from various aquatic, aerophytic, and synanthrophic substrates from Europe, America and Oceania. The other two investigated strains M3 and D1 are closely related with high support with lineage E4 (Škaloud and Rindi 2013). Members of this lineage were reported so far from different aerophytic, rock, soil ,and synanthrophic substrates from Europe and Australia.
Phylogenetic tree based on Bayesian analysis of combined newly retrieved rbcL sequences and sequences obtained from GenBank, showing the position of investigated Klebsormidium strains (bold) and related genus Interfilum. Values at nodes indicate statistical support estimated by MrBayes posterior node probability (left), maximum likelihood bootstrap (middle), and maximum parsimony bootstrap (right). Thick branches represent nodes with highest posterior node probability (1.00). Superclade (A–G) and clade designation (E1–E5) followed Rindi et al. (2011). Numbers 1–13 give strains affiliation to 13 lineages sensu Škaloud and Rindi (2013). Scale bar indicates numbers of substitutions per site
Photosynthesis under light gradient
The relative electron transport rate as a function of increasing photon fluence rates up to 314 μmol photons m−2 s−1 differed between the strains investigated (Fig. 3a, b). Strains of lineage E2 exhibited α values of 0.475 in M1, 0.525 in G6, and 0.475 in G7 (Fig. 3a). While the relative ETRmax was highest for M1 (25.8), G6 and G7 had lower rates with 21.6 and 19.3, respectively (Fig. 3a). The lk value was 55 μmol photons m−2 s−1 for M1 and 40 μmol photons m−2 s−1 for G6, which are higher compared to G7 that exhibited an lk value of 19 μmol photons m−2 s−1 (Fig. 3a). Slight photoinhibition in response to increasing photon fluence densities up to 314 μmol photons m−2 s−1 could be observed in strains M1 and G6. Both E4 lineage strains showed α values of 0.231 and 0.236, which were significantly lower in comparison with those of the E2 lineage strains (p ≤ 0.001; Fig 3b). The relative ETRmax was lower as well (14.1 and 15.7), and lk was higher than in E2 lineage strains (63 and 68 μmol photons m−2 s−1; Fig 3b). However, in both E2 lineage strains M3 and D1, there was no indication of photoinhibition (Fig. 3b).
Relative electron transport rate (rETR) curves in response to increasing photon fluence densities up to 314 μmol photons m−2 s−1 (n = 4, mean ± SD). a Strains of lineage E2: Klebsormidium sp. M1, Klebsormidium sp. G6, and Klebsormidium sp. G7. b Strains of lineage E4: Klebsormidium sp. M3 and Klebsormidium sp. D1. Data points were fitted according to Walsby (1997)
Growth under temperature and light gradient
The effect of increasing temperatures showed the capability for growth from 18.1 to 27.9 °C for all investigated Klebsormidium strains, except strain D1, which exhibited only a low growth rate (≤0.2 μ−1 day−1) at 26.3 °C and at the highest tested temperature of 27.9 °C (Fig. 4). The optimum growth temperature for lineage E2 strains varied between 18.1 and 24.6 °C, with growth rates between 0.35 and 0.42 μ day−1 (Fig. 4). Strain G7 (lineage E2) showed a surprising pattern, with the highest growth rate of 0.42 μ day−1 at 18.1 °C and at 26.3 °C with 0.43 μ day−1 slightly higher, whereas between 19.1 and 24.6 °C, the growth rate was lower (0.27–0.38 μ day−1). Strains of Klebsormidium lineage E4 had the highest growth rates with 0.42–0.43 μ day−1 at 19.1 °C (M3) and 18.1 °C (D1) (Fig. 4). There were no significant differences of optimum growth temperature between strains of lineage E2 and E4.
Growth rate (μ day−1) of the five investigated Klebsormidium strains collected from BSCs at the semi-arid Colorado Plateau under six different temperatures, ranging from 18.1 to 27.9 °C measured at 40 μmol photons m−2 s−1 and a light/dark cycle of 16:8 h L/D (mean, n = 4). Asterisks mark highest measured growth rate under tested conditions. Additionally, clade assignment followed Rindi et al. (2011) for each strain is given
The effect of increasing photon fluence rates showed the capability for growth from 4 to 110 μmol photons m−2 s−1 for all five investigated Klebsormidium strains (Fig. 5). All strains exhibited their highest grow rates at medium to highest tested photon fluence rates. At 4 μmol photons m−2 s−1, all strains grew only poorly, with growth rates between 0.11 and 0.19 μ day−1 (Fig. 5). Strains of E2 lineage (M1, G6, and G7) exhibited the highest growth rate at 30–110 μmol photons m−2 s−1 (Fig. 5). Strain M1 showed the highest growth rate of 0.37 μ day−1 at 110 μmol photons m−1 s−1, whereas the growth rates at 30 and 85 μmol photons m−2 s−1 with 0.32 and 0.35 μ day−1 were quite similar (p > 0.001; Fig. 5). Consequently, strain M1 exhibited no indication of inhibition under increasing photon fluence densities. For strain G6, an increase of photon fluence densities up to 85 μmol photons m−2 s−1 was accompanied by an almost linear rise in the growth rate up to a maximum of 0.34 μ day−1 (p > 0.01; Fig. 5). Exposure to 110 μmol photons m−2 s−1 led to a decrease of the growth rate down to 0.28 μ day−1 (Fig. 5). Strain G7 exhibited a similar response pattern, with the highest growth rate of 0.40 μ day−1 at already 30 μmol photons m−2 s−1 (p > 0.01; Fig. 5). The increase in photon fluence densities up to 110 μmol photons m−2 s−1 led first to a slight and then a stronger decrease of the growth rate (0.35 and 0.28 μ day−1, respectively; Fig. 5). G6 and G7 exhibited low light requirements for growth. For both lineage E4 strains (M3 and D1), the increase of photon fluence densities up to 85 μmol photons m−2 s−1 were accompanied by an almost logarithmic rise in the growth rate up to a maximum of 0.44 and 0.38 μ day−1, respectively (p > 0.001; Fig. 5). The further increase of photon fluence densities up to 110 μmol photons m−2 s−1 resulted for strain D1 a slight reduction in growth rates (0.35 μ day−1), while in strain M3, the growth remained unchanged (0.44 μ day−1). Both strains showed no indication of inhibition under increasing photon fluence densities. There were no significant differences in the growth rates as function of the tested photon fluence rates between strains of lineage E2 and E4.
Growth rate (μ day−1) of the five investigated Klebsormidium strains collected from BSCs at the semi-arid Colorado Plateau under increasing photon fluence densities, ranging from 4 to 110 μmol photons m−2 s−1 measured at 20 °C (±1 °C) and a light/dark cycle of 16:8 h L/D (mean, n = 4). The significance of differences among photon fluence densities are indicated by different letters (small letters for strain M1; capital letters for strain M3; cursive small letters for strain D1; cursive capital letters for strain G6; bold small letters for strain G7) and was calculated by one-way ANOVA (p < 0.001 for M1, M3, and D1; p < 0.01 for G6 and G7). Asterisks mark highest measured growth rate under tested conditions. Additionally, clade assignment that followed Rindi et al. (2011) for each strain is given
Desiccation and rehydration
The standardized methodological approach with the dry chamber and PAM measurements from outside allowed comparative effective quantum yield determinations in all samples during the desiccation experiment. All tested Klebsormidium strains gave a positive signal of ΔF/fm′ for at least 370 min, before a threshold was reached, after which the signal rapidly decreased to a quantum yield of zero or values near to zero (data not shown). The three strains M1, G6, and G7 assigned to lineage E2 gave a positive fluorescence signal for 390 to 430 min, whereas the two strains M3 and D1 of lineage E4 exhibited a positive signal for a slightly shorter time interval (370 and 395 min). Two representative examples for E2 (strain M1) and E4 (D1) are shown in Fig. 6.
The effect of controlled desiccation and rehydration on the effective quantum yield (ΔF/Fm′) of photosystem II in 2 representatives out of 5 Klebsormidium strains collected from BSCs at the semi-arid Colorado Plateau. Strains M1 and D1 are representative for clade E2 and E4, respectively. ΔF/Fm′ was measured at 30–40 μmol photons m−2 s−1 with a PAM 2500 according the approach of Karsten et al. (2014) (n = 4, mean value ± SD)
After the signal of quantum yield reached zero or values near to zero, the samples were rehydrated and all samples recovered within 90 min, at least partially (data not shown). The recovery of the effective quantum yield was highly variable in Klebsormidium strains of lineage E2. The recovery in strain M1 was 32.9% (Fig. 6), in G6 86.7% and in G7 66.3%. In contrast, both E4 lineage strains showed with 95.4% (M3) and 97.6% (D1) (Fig. 6), an almost complete recovery. The differences of recovery rates between strains of lineage E2 and E4 were highly significant (p ≤ 0.01).
Discussion
Molecular analyses
Škaloud and Rindi (2013) proposed within Klebsormidium superclade E the existence of three different habitat-type preferences (natural subaerial, artificial subaerial, and aquatic) and hence distinct genotypes and ecotypes. Some of the described lineages seem to consist exclusively of strains with one habitat-type preference; others compromised strains with different habitat types (E4 and E2, respectively). As the present results on strains M1, G6, and G7 grouped to the original habitat-type preference circumcising exclusively “artificial,” our data contradict partially the assumed ecology of the Klebsormidium superclade E. This can be caused by two reasons: (I) either previous studies were not sufficiently comprehensive and hence not representative simply by not sampling certain biogeographical regions or (II) the sole use of the genetic marker rbcL failed to display the real genetic diversity. Concerning the first case, the focus of previous studies was indeed mainly Europe (e.g., Škaloud and Rindi 2013). Strains from northwestern American dryland BSCs were not included so far in earlier studies. Concerning the second case, it is known that analysis based on single genes sometimes fail to distinguish between closely related Klebsormidium strains (e.g., Mikhailyuk et al. 2015). A combination of ITS rDNA and rbcL gene would give higher nodal support and seems to be more suitable for the detection of cryptic species (Rindi et al. 2011; Škaloud and Rindi 2013).
The most common genotypes in Klebsormidium worldwide are members of superclade B/C and E (Mikhailyuk et al. 2015; Ryšánek et al. 2015). In many habitats, such as alpine sites in Tyrol (Austrian Alps), at least these two B/C and E genotypes dominate, although also others (e.g., superclade F) have been reported (Mikhailyuk et al. 2015). In contrast, BSCs of the Colorado Plateau harbor only members of the superclade E. Only few other locations seem to be solely dominated by the Klebsormidium E clade, such as algal biofilms on concrete, bark of trees, and rocks in Washington, Ohio, and Connecticut, USA (Ryšánek et al. 2015). Until now, the present investigation is the first study on the biodiversity of Klebsormidium strains from BSCs in North American drylands. So far, and as there are no other studies, it is impossible to make far-reaching conclusions on the genetic diversity of Klebsormidium in BSCs of the Colorado Plateau.
Photosynthesis under a light gradient
The effect of increasing photon fluence densities on rETR revealed only minor differences among the five Klebsormidium isolates. All strains exhibited a relatively high α together with low values of lk, although a small but significant difference in α between E2 and E4 members was noted. The combination of a high α together with low lk value is typical for low-light adapted algae (Henley 1993). Such low-light response patterns are usually coupled with photoinhibition under elevated irradiance levels (Bischof et al. 1998). Photoinhibition could not be statistically documented in the present study, although in some strains, at least such a trend is visible (Fig. 3). The tested Klebsormidium strains rather confirm previous studies on other Klebsormidium representatives and related genera Interfilum, Entransia, and Hormidiella (Karsten et al. 2010, 2014, 2016; Karsten and Holzinger 2012; Aigner et al. 2013; Holzinger and Karsten 2013), which all document lack of photoinhibition at least under 2–3-fold higher photon fluence rates compared to the present study. These results were interpreted as high photophysiological plasticity of the mentioned algal genera. The herein observed slightly photoinhibition of strains M1 and G6, as well the lower Pmax and higher lk values of D1 and M3 in comparison with the other three strains, might be the result of different small-scale spatial distribution of Klebsormidium filaments and strains within the BSCs. Depending on the succession stage of BSCs and water availability, BSC species composition changes and hence vertical distribution of crust members (Garcia-Pichel et al. 2016). The vertical structure of BSCs results in the occupation of microenvironments within the BSC where individual cells are protected from damaging light levels (Gray et al. 2007). A more recent study on the three-dimensional structure of BSCs showed a steep decline in irradiance with depth, but also the existence of “light pockets” where the irradiance was 10-fold higher than in adjacent parts at the same depth (Raanan et al. 2016). The photophysiological plasticity of Klebsormidium might allow the occurrence along and exploitation of different vertical micro-environmental gradients (e.g., solar radiation) in the complex three-dimensional BSC structure, including light pockets (see above). The advantage of utilization of different light levels within the crust is a protection from damaging irradiance levels (Gray et al. 2007). Furthermore, Klebsormidium might be protected through multi-layered carpet-like structures, which contribute to self-shading and protection of single filaments (Karsten et al. 2010). All these data indicate that even in high insolation habitats such as the Colorado Plateau, BSCs provide a protective environment for many microorganisms due the specific spatial and physicochemical structure.
Growth under a temperature and light gradient
The effect of increasing temperatures on growth indicated the capability of all strains to cope with the tested temperature range. All Klebsormidium strains were able to grow at the highest average air temperature of the sampling sites (25 °C in Moab, 25.7 °C Dead Horse Point, 20 °C Grand Canyon; Table 1). The optimum growth temperature was relatively high for all strains, and hence, they can be regarded as psychrotolerant microorganisms (Elster 1999). As the herein tested maximum temperature was 27.9 °C because of methodological limitations, it is not possible to predict if the five strains are able to grow also at higher temperatures. The same is true for temperatures under 18.1 °C, as they were also not tested. Nevertheless, the growth-temperature response patterns of all investigated Klebsormidium strains well reflect the seasonally occurring temperature fluctuations in the Colorado Plateau habitats.
The effect of increasing photon fluence densities on growth in all the five tested strains showed an optimum between 30 and 110 μmol photons m−2 s−1. Two strains (G6 and G7) showed at 85 and 110 μmol photons m−2 s−1, respectively, already growth inhibition and hence low light requirements for growth. In contrast, in the remaining three strains, higher photon fluence densities did not lead to a reduction in growth rates. This is an interesting result, because the few existing studies on different Klebsormidium species pointed always to low light requirements for photosynthesis and growth. K. crenulatum, K. dissectum, and a no further specified Klebsormidium species exhibited their optimum growth at 30 μmol photons m−2 s−1, along with always a strong reduction in growth rates at further increasing photon fluence densities (Karsten and Rindi 2010; Karsten et al. 2010; Karsten and Holzinger 2012). Other terrestrial filamentous green algae of the genus Trentepohlia, such as Trentepohlia odorata and T. aurea, which grow on tree barks and building surfaces, showed maximum growth rates between ∼60 and 120 μmol photons m−2 s−1 (Tan et al. 1993; Abe et al. 1998). Such growth data were always interpreted as an adaption to low light conditions, which are in agreement with the low-light requirements for photosynthesis (see above) (Karsten et al. 2010). Besides specific micro-habitat properties that support light gradients inside a BSC, there exist various physiological mechanisms to avoid photoinhibition or even photodamage in algae. These include, for example, phototactic movement of plastids within cells, minimization of absorption, and utilization of incident PAR for photosynthesis (Raven 2011). Such processes aim to dissipate enhanced excitation of photosynthetic pigments, for example, by non-photochemical quenching (xanthophyll cycle) and photochemical quenching (alternative electron transport pathways) (Raven 2011). The underlying genes for such protective non-photochemical quenching, i.e., cyclic electron flow activity at photosystem I, have recently been found in the genome of K. flaccidum (Hori et al. 2014).
Dehydration and rehydration
The dehydration of the five tested Klebsormidium strains of superclade E under standardized conditions resulted in an inhibition of the effective quantum yield of PSII. All strains gave a positive fluorescence signal for 370 to 430 min of exposure to ∼10% relative air humidity, with no statistical difference between members of lineage E2 and E4. A recent study including nine E-clade strains from temperate BSCs in Germany resulted with 350–480 min in a similar time period until the effective quantum yield was completely inhibited (Donner et al. 2016), while a clade E6 Klebsormidium strain isolated from a snow layer in Alaska exhibited with 300 min a much shorter positive photosynthetic signal (Karsten et al. 2016). However, few ecophysiological data exist on desiccation of E-clade strains originating from other geographic regions. The present results on the 5 E-clade strains of Klebsormidium from the Colorado Plateau indicate that there is no differentiation between genotypes possibly based on the dehydration kinetics only.
In strong contrast, the recovery kinetics of ΔF/fm′ after dehydration of the desiccated Klebsormidium strains exhibited lineage-specific differences. Strains of lineage E2 exhibited a large variability in their recovery kinetics leading to 33–87% of the control. The two E4 lineage strains M3 and D1 showed a complete recovery of the control effective quantum yield. Similar recovery kinetics have been observed in E-clade strains from Alaska (Karsten et al. 2016), as well as in closely related members of the genera Interfilum, Entransia, and Hormidiella with recovery rates between 70 and 100% (Karsten et al. 2014; Herburger and Holzinger 2015). High recovery rates seem to be a specific trait of terrestrial green algae, as numerous Chlorophyta show the same high recovery rates (Gray et al. 2007). In contrast, aquatic Chlorophyta typically exhibit much lower recovery rates after desiccation (e.g., Pichrtová et al. 2014). The present study suggests lineage- and hence genotype-specific differences of the recovery kinetics. Such strain-specific differences in desiccation tolerance have already been observed in a recent study on different Klebsormidium species and strains of superclades B/C and E (Donner et al. 2016).
Mikhailyuk et al. (2015) provided a concept, in which E clade strains are considered as typically adapted to more hydrophilic habitats, compared, for example, to the xerophytic F clade. This ecological concept is based on ecophysiological data and a wide distribution of E clade members in humid and shaded habitats of Southwestern Europe, Asia, and North America (Mikhailyuk et al. 2015, and references therein). The present phylogenetic and ecophysiological results of E-clade strains from North American dryland BSCs contradict this assumption. The physiological properties reported in the present study rather indicate xerophytic traits in the Klebsormidium E-clade strains from the Colorado Plateau. The environmental conditions at all three sampling sites on the Colorado Plateau are semi-arid, with low monthly (0–50 mm) and annual precipitation (200–390 mm), and hence, these data well support the xerophytic features of the investigated five E-clade strains of Klebsormidium.
Conclusions
In the present study, only clade E members of Klebsormidium were found in BSCs collected at the semi-arid Colorado Plateau. Until now, clade E members have mainly be reported in humid habitats in southwestern Europe, Asia, and North America (Mikhailyuk et al. 2015), and hence, the biogeography of clade E has to be revised concerning drylands. The ecophysiological response patterns as function of desiccation, temperature, and light indicate a pronounced plasticity within members of the genus Klebsormidium. All data well explain the ecological success of these green algae in BSCs of the Colorado Plateau.
References
Abe K, Mihara H, Hirano M (1998) Characteristics of growth and carotenoid accumulation of the aerial microalga Trentepohlia aurea in liquid culture. J Mar Biotech 6:53–58
Aigner S, Remias D, Karsten U, Holzinger A (2013) Unusual phenolic compounds contribute to ecophysiological performance in the purple-colored green alga Zygogonium ericetorum (Zygnematophyceae, Streptophyta) from a high-alpine habitat. J Phycol 49:648–660
Belnap J (1994) Potential role of cryptobiotic soil crust in semiarid rangelands. In: Monsen SB, Kitchen SG (eds) Proceedings: symposium on ecology, management, and restoration of intermountain annual rangelands. U.S. Department of Agriculture, National Forest Service, Intermountain Research Station, Fort Collins, pp 179–185
Belnap J (2002) Nitrogen fixation in biological soil crusts from southeast Utah, USA. Biol Fertil Soils 35:128–135
Belnap J (2003) Comparative structure of physical and biological soil crust. In: Belnap J, Lange OL (eds) Biological soil crusts: structure, function, and management. Springer, Berlin, pp 177–191
Belnap, Weber B, Büdel B (2016) Biological soil crusts as an organizing principle in drylands. In: Weber B, Büdel B, Belnap J (eds) Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, pp 3–13
Bischof K, Hanelt D, Tüg H, Karsten U, PEM B, Wiencke C (1998) Acclimation of brown algal photosynthesis to ultraviolet radiation in Arctic coastal waters (Spitsbergen, Norway). Polar Biol 20:388–395
Büdel B, Dulic T, Darienko T, Rybalka N, Friedl T (2016) Cyanobacteria and algae of biological soil crusts. In: Weber B, Büdel B, Belnap J (eds) Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, pp 55–80
Donner A, Glaser K, Borchhardt N, Karsten U (2016) Ecophysiological response on dehydration and temperature in terrestrial Klebsormidium (Streptophyta) isolated from biological soil crusts in Central European grasslands and forests. Microb Ecol DOI: 10.1007/s00248-016-0917-3.
Elbert W, Weber B, Burrows S, Steinkamp J, Büdel B, Andreae MO, Pöschl U (2012) Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nat Geosci 5:459–462
Elster J (1999) Algal versatility in various extreme environments. In: Seckbach J (ed) Enigmatic microorganisms and life in extreme environments. Springer, Dordrecht, pp 215–227
Garcia-Pichel F, Felde V, Drahorad S, Weber B (2016) Microstructure and weathering processes within biological soil crusts. In: Weber B, Büdel B, Belnap J (eds) Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, pp 237–255
Glaser K, Donner A, Albrecht M, Mikhailyuk T, Karsten U (2017) Habitat-specific composition of morphotypes with low genetic diversity in the green algal genus Klebsormidium (Streptophyta) isolated from biological soil crusts in Central European grasslands and forests. Eur J Phycol, DOI: 10.1080/09670262.2016.1235730
Gray DW, Lewis LA, Cardon ZG (2007) Photosynthetic recovery following desiccation of desert green algae (Chlorophyta) and their aquatic relatives. Plant Cell Environ 30:1240–1255
Gustavs L, Schumann R, Eggert A, Karsten U (2009) In vivo growth fluorometry: accuracy and limits of microalgal growth rate measurements in ecophysiological investigations. Aquat Microb Ecol 55:95–104
Henley WJ (1993) Measurement and interpretation of photosynthetic light-response curves in algae in the context of photoinhibition and diel changes. J Phycol 29:729–739
Hepperle D (2004) SeqAssem. Version: Sequentix. A sequence analysis tool, Contig assembler.
Herburger K, Holzinger A (2015) Localization and quantification of callose in the Streptophyte green algae Zygnema and Klebsormidium: correlation with desiccation tolerance. Plant Cell Physiol 56:2259–2270
Holzinger A, Karsten U (2013) Desiccation stress and tolerance in green algae: consequences for ultrastructure, physiological and molecular mechanisms. Front Plant Sci 4:327
Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M et al (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5:1–9
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinform 17:754–755
Karsten U, Lütz C, Holzinger A (2010) Ecophysiological performance of the aeroterrestrial green alga Klebsormidium crenulatum (Charophyceae, Streptophyta) isolated from an alpine soil crust with an emphasis on desiccation stress. J Phycol 46:1187–1197
Karsten U, Rindi F (2010) Ecophysiological performance of an urban strain of the aeroterrestrial green alga Klebsormidium sp. (Klebsormidiales, Klebsormidiophyceae). Eur J Phycol 45:426–435
Karsten U, Holzinger A (2012) Light, temperature, and desiccation effects on photosynthetic activity, and drought-induced ultrastructural changes in the green alga Klebsormidium dissectum (Streptophyta) from a high alpine soil crust. Microb Ecol 63:51–63
Karsten U, Herburger K, Holzinger A (2014) Dehydration, temperature and light tolerance in members of the aeroterrestrial green algal genus Interfilum (Streptophyta) from biogeographically different temperate soils. J Phycol 50:804–816
Karsten U, Herburger K, Holzinger A (2016) Living in biological soil crust communities of African deserts—physiological traits of green algal Klebsormidium species (Streptophyta) to cope with desiccation, light and temperature gradients. J Plant Physiol 194:2–12
Lokhorst GM (1996) Comparative taxonomic studies on the genus Klebsormidium (Charophyceae) in Europe. Cryptogamic studies 5. Gustav Fischer Verlag, Stuttgart, p 132
Mikhailyuk T, Holzinger A, Massalski A, Karsten U (2014) Morphological and ultrastructural aspects of phylogenetically closely related genera Interfilum and Klebsormidium (Klebsormidiales, Streptophyta). Eur J Phycol 49:395–412
Mikhailyuk T, Glaser K, Holzinger A, Karsten U (2015) Biodiversity of Klebsormidium (Streptophyta) from alpine biological soil crusts (Alps, Tyrol, Austria, and Italy). J Phycol 51:750–767
Nylander JA (2004) MrModeltest v2. Version. Evolutionary Biology Centre, Uppsala University, Sweden
Pichrtová M, Hájek T, Elster J (2014) Osmotic stress and recovery in field populations of Zygnema sp. (Zygnematophyceae, Streptophyta) on Svalbard (high Arctic) subjected to natural desiccation. FEMS Microb Ecol 89:270–280
R Development Core Team (2009) R. A language and environment for statistical computing. Version: R foundation for statistical computing (http://www.R-project.org).
Raanan H, Felde V, Peth S, Drahorad S, Ionescu D, Gil E et al (2016) Three-dimensional structure and cyanobacterial activity within a desert biological soil crust. Environ Microbiol 18:372–383
Raven JA (2011) The cost of photoinhibition. Physiol Plant 142:87–104
Rindi F, Mikhailyuk T, Sluiman HJ, Friedl T, Lopez-Bautista JM (2011) Phylogenetic relationships in Interfilum and Klebsormidium (Klebsormidiophyceae, Streptophyta). Mol Phylogenet Evol 58:218–231
Rosentreter R, Belnap J (2003) Biological soil crusts of North Amercia. In: Belnap J, Lange OL (eds) Biological soil crusts: structure, function, and management. Springer, Berlin, pp 31–50
Ryšánek D, Hrčková K, Škaloud P (2015) Global ubiquity and local endemism of free-living terrestrial protists: phylogeographic assessment of the streptophyte alga Klebsormidium. Environ Microbiol 17:689–698
Schreiber U, Bilger W (1993) Progress in chlorophyll fluorescence research: major developments during the past years in retrospect. In: Behnke HD, Lüttge U, Esser K, Kadereit JW, Runge M (eds) Progress in botany. Springer, Berlin, pp 151–173
Škaloud P, Lukesová A, Malavasi V, Ryšánek D, Hrcková K, Rindi F (2014) Molecular evidence for the polyphyletic origin of low pH adaptation in the genus Klebsormidium (Klebsormidiophyceae, Streptophyta). Plant Ecol Evol 147:333–345
Škaloud P, Rindi F (2013) Ecological differentiation of cryptic species within an asexual protist morphospecies: a case study of filamentous green alga Klebsormidium (Streptophyta). J Eukaryot Microb 60:350–362
Starr RC, Zeikus JA (1993) UTEX—the culture collection of algae at the University of Texas at Austin 1993 list of cultures. J Phycol 29:1–106
Swofford DL (2002) PAUP* phylogenetic analysis using parsimony (∗and other methods). Version 4. Version 4. Sinauer Associates, Sunderland
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tan CK, Lee YK, Ho KK (1993) Effect of light intensity and ammonium-N on carotenogenesis of Trentepohlia odorata and Dunaliella bardawil. J Appl Phycol 5:547–549
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Walsby AE (1997) Numerical integration of phytoplankton photosynthesis through time and depth in a water column. New Phytol 136:189–209
Williams L, Borchhardt N, Colesie C, Baum C, Komsic-Buchmann K, Rippin M et al (2017) Biological soil crusts of Arctic Svalbard and of Livingston Island, Antarctica. Polar Biol 40:399–411
Woelfel J, Schoknecht A, Schumann R, Karsten, U (2014) Growth and primary production characteristics of three benthic diatoms from the brackish Southern Baltic Sea in relation to varying environmental conditions. Phycologia 53:639–651
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin, USA
Acknowledgments
We thank Dr. Karin Glaser (University of Rostock) for support with the statistical data analysis. DR was supported by Charles University Science Foundation [GAUK no. 1544214], TM thanks the Alexander von Humboldt Foundation for financial support in the frame of a Georg-Forster-Fellowship, and UK greatly appreciates funding by the German Research Council (DFG) (KA899/16).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Donner, A., Ryšánek, D., Mikhailyuk, T. et al. Ecophysiological traits of various genotypes of a green key alga in biological soil crusts from the semi-arid Colorado Plateau, USA. J Appl Phycol 29, 2911–2923 (2017). https://doi.org/10.1007/s10811-017-1158-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-017-1158-7