Abstract
Reduced corpus callosum area and increased brain volume are two commonly reported findings in autism spectrum disorder (ASD). We investigated these two correlates in ASD and healthy controls using T1-weighted MRI scans from the Autism Brain Imaging Data Exchange (ABIDE). Automated methods were used to segment the corpus callosum and intracranial region. No difference in the corpus callosum area was found between ASD participants and healthy controls (ASD 598.53 ± 109 mm2; control 596.82 ± 102 mm2; p = 0.76). The ASD participants had increased intracranial volume (ASD 1,508,596 ± 170,505 mm3; control 1,482,732 ± 150,873.5 mm3; p = 0.042). No evidence was found for overall ASD differences in the corpus callosum subregions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Two commonly reported neuroanatomical correlates of autism are reduced corpus callosum area and increased brain volume (Frazier and Hardan 2009). We investigated corpus callosum area and brain volume differences in autism spectrum disorder (ASD) using structural MRI scans provided with the recently released Autism Brain Imaging Data Exchange (ABIDE) database, a large multi-site public neuroimaging dataset of individuals with autism and healthy controls (Di Martino et al. 2013).
There are some notable differences between this study and previous investigations of corpus callosum area and brain volume in autism. The ABIDE dataset consists of MRI scans of 1112 participants (autism and controls), which is considerably larger than the sample sizes of previous studies that investigated ASD using structural MRI. For example, typical sample sizes for corpus callosum studies range from 20 to 102 (total n, patients + controls) (Egaas et al. 1995; Rice et al. 2005). The high number of subjects in the ABIDE dataset will allow us to detect smaller differences than previous studies. Participants in the ABIDE study were recruited and imaged at multiple centers, increasing the generalizability of this analysis for the autism population that was sampled in ABIDE.
Corpus callosum mid-sagittal area and intracranial volume were measured using automated quantitative software methods and analyses were carried out blind to participant status. We investigated changes in corpus callosum area and intracranial volume in autism spectrum disorders, and additionally investigated differences in these measures in ASD subtypes, defined according to DSM-IV criteria. Regional corpus callosum changes were further investigated by subdividing the corpus callosum using the Witelson scheme (Witelson 1989).
Specific hypotheses are listed below. Age, sex and site were included as covariates in all statistical models. Intracranial volume was included as a covariate for corpus callosum area analyses.
-
1.
Corpus callosum mid-sagittal cross-sectional area is different in ASD and ASD subgroups relative to healthy controls.
-
2.
Intracranial volume is different in ASD and ASD subgroups relative to healthy controls.
-
3.
Corpus callosum subregional areas are different in ASD and ASD subgroups relative to healthy controls.
All corpus callosum segmentations, brain segmentations, quantitative measurements and scripts for statistical analyses and plots will be provided for download at https://sites.google.com/site/hpardoe/cc_abide.
Materials and Methods
Subject Recruitment and MR Image Acquisition
Structural MRI data from the Autism Brain Imaging Data Exchange (ABIDE) database were used for these analyses. A total of 1112 participants were included (539 autism cases and 573 healthy controls). No identifying information were provided with the MRI scans in accordance with HIPAA guidelines. Each institution’s human subjects research board established the criteria of informed consent. Further information about the study can be found at the ABIDE website (http://fcon_1000.projects.nitrc.org/indi/abide/). Corpus callosum area and intracranial volume were assessed using whole brain T1-weighted MRI acquired as part of the ABIDE study. Image acquisition parameters and number of autism cases and controls for each site are provided in Table 1.
Image Processing
Quality Assessment
Two methods were used to assess the quality of each structural MRI scan. The first method was a qualitative visual assessment of motion artifact. Each MRI scan was visually inspected for motion artifact and rated from 1 to 5, with 1 indicating the most severe motion artifact and 5 indicating images free from any detectable motion-related artifact (see Supplementary data for example image ratings). The second QA procedure was a quantitative, objective method that used resting state fMRI data, which were also acquired for each participant as part of ABIDE, to estimate the participant’s motion during the scan. The “MCFLIRT” software tool provided as part of FSL was used to estimate an affine transform for each volume acquired as part of the resting state scan to a common volume, and the root mean square deviation between consecutive volumes was estimated using the tool “rmsdiff” (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/MCFLIRT, (Jenkinson et al. 2002). These values were averaged over the entire acquisition to obtain an estimate of the amount of movement during the resting state fMRI. The average root mean square deviation was used as a proxy measure of motion for each participant.
The primary analyses excluded any scans with a qualitative rating of 1 or 2. We repeated the analyses with the resting state fMRI-derived quantitative motion measure included as a covariate to determine if this objective measure had any impact on our findings.
Corpus Callosum Area and Intracranial Volume Measurement
The corpus callosum for each individual was segmented using “yuki” (Ardekani 2013). In addition to segmentation of the entire corpus callosum in the mid-sagittal plane, the corpus callosum was automatically subdivided using the Witelson scheme (Witelson 1989). Corpus callosum segmentations were visually inspected and if necessary minor corrections were carried out using the software package “ITK-SNAP” (Yushkevich et al. 2006). Two readers carried out the visual inspection and correction procedure (RKH and RA).
Since corpus callosum segmentations involved minor manual corrections for some scans, the results could be operator-dependent. All scans from one site (Michigan dataset #1) were segmented by both readers and a statistical equivalence analysis was carried out [α = 0.05, equivalence margin = 5 %, (Robinson 2010)] to determine if corpus callosum segmentations carried out by each reader could be pooled. Intra-class correlation was also calculated using corpus callosum areas estimated by both readers.
Total intracranial volume was estimated using the software tool “brainwash”, distributed as part of the software package “Automatic Registration Toolbox” (www.nitrc.org/projects/art). The brainwash method uses non-linear registration of a number of MRI scans that have pre-labeled intracranial regions to the participant’s MRI scan that we wish to estimate the intracranial volume. The coregistered labels are then mapped to the participant’s brain using the estimated non-linear transform. A voxel-wise voting scheme is then used to classify each voxel in the participant MRI as intracranial or not. All brain segmentations were visually inspected to ensure accurate segmentation. In some cases where segmentation was inaccurate, the following additional processing steps were used in order:
-
1.
Site specific atlases: If inaccurate brain segmentations were identified, the brainwash procedure was rerun using processed MRI scans from the same site that had error-free brain segmentations.
-
2.
Correcting for inaccurate anterior commissure and posterior commissure coordinates: The brainwash software relies on automatic detection of the anterior and posterior commissures on the mid-sagittal plane. In some cases these landmarks were incorrectly identified. In these cases the anterior and posterior commissure were manually identified and entered into brainwash.
-
3.
Manual brain correction: Further minor intracranial volume segmentation errors were corrected manually using the software package ITK-SNAP (Yushkevich et al. 2006). A few iterations of the automated segmentation method “region-based snakes” implemented in ITK-SNAP were often useful for obtaining accurate intracranial volume segmentations.
All edits to corpus callosum segmentations or intracranial volume were carried out blind to subject diagnosis.
Statistical Analysis
All analyses of corpus callosum area (total area and Witelson subdivision areas) between ASD cases and controls were investigated using a general linear model, with the corpus callosum area as the dependent variable and diagnosis, age, sex, intracranial volume and site as the independent variables. Age was modeled as a quadratic function. Interaction terms between age and diagnosis, and intracranial volume and diagnosis were included in the models. For the investigation of intracranial volume differences between the ASD and control groups, intracranial volume was the dependent variable and the other factors described above as independent variables.
The same model parameters were used for ASD subgroups. All model specifications are provided at https://sites.google.com/site/hpardoe/cc_abide, implemented in the R software package (R Core Team 2013).
Throughout the text effect sizes are reported as a percentage difference relative to healthy controls. Cohen’s d was also calculated for the primary hypothesis tested in this study (overall corpus callosum area changes in ASD) in order to compare with a previous meta-analysis (Frazier and Hardan 2009), and any other relevant findings.
Results
The ABIDE dataset contained 1112 participants. From this dataset, corpus callosum area measurements were obtained for 1100 participants. An example corpus callosum segmentation is shown in Fig. 1. Corpus callosum area measurements were not obtained in 12 cases for the following reasons: One participant’s structural MRI scan had severe motion artifact causing yuki to fail, ten participants did not have a structural MRI scan provided for download, and one participant’s structural MRI scan was unable to be processed using yuki due to enlarged ventricles. The 1100 subjects consisted of 569 healthy controls and 531 participants with ASD. Sixty five participants (45 autism and 20 controls) were graded as having motion artifact (visual assessment scores of 1 and 2) and were excluded from the final analysis, leaving 1035 participants total, comprising 486 autism cases and 549 controls. ASD participants were further classified according to DSM-IV criteria as Autism (n = 315), Asperger’s syndrome (n = 86), Pervasive Developmental Disorder not otherwise specified (PDD-NOS, n = 32) and Asperger’s or PDD-NOS (n = 6). Classifications for 47 ASD participants were not provided by ABIDE. Further demographic information regarding these ASD DSM-IV classifications is outlined in Table 2.
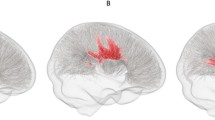
Quantitative assessment of corpus callosum area in autism spectrum disorders. The figure shows example data from an ASD participant in the ABIDE study. a The midsagittal plane is generated from a 3D volumetric T1 weighted MRI scan. b Corpus callosum segmentation is shown in red. The area enclosed by the red line is measured. c The corpus callosum is further divided according to the Witelson scheme. Region W1 (rostrum) is shown in red, W2 (genu) in orange, W3 (anterior body) in yellow, W4 (mid-body) in green, W5 (posterior body) in blue, W6 (isthmus) in purple and W7 (splenium) in light purple. Corpus callosum and brain segmentation data are provided at https://sites.google.com/site/hpardoe/cc_abide (Color figure online)
Minor manual editing was performed using ITK-SNAP software on 38.9 % of MRI scans. The percentage of segmentation edits per site ranged from 0 to 96.2 %. Corpus callosum measures from the two readers had an ICC = 0.94 and were found to be statistically equivalent (equivalence margin 5 %, p = 1.3 × 10−14).
Corpus Callosum Midsagittal Area in ASD and DSM-IV Subgroups
There was no evidence for differences in corpus callosum area between ASD participants and controls (Fig. 2, ASD 598.53 ± 109 mm2; control 596.82 ± 102 mm2; Cohen’s d = 0.016; p = 0.76). Including the fMRI-derived motion estimate as a covariate did not change our findings; no difference between control and ASD participants was detected (p = 0.34), and no significant correlation between participant motion and corpus callosum area was observed (p = 0.35). A plot of corpus callosum area as a function of age is shown in Fig. 2, indicating that there are no distinguishable differences in corpus callosum area in individuals with ASD compared with healthy controls, over the age range of participants in the ABIDE study. A subtle reduction in corpus callosum area was observed in the Autism DSM-IV subtype (−13.42 mm2, p = 0.045). No evidence for corpus callosum area reduction was found in the Asperger’s or PDD-NOS category (Asperger’s p = 0.69; PDD-NOS p = 0.88). Differences in the Asperger’s or PDD-NOS category approached significance (p = 0.088), however this group only contained 6 participants.
No difference was found in corpus callosum mid-sagittal area between ABIDE ASD participants and controls. Dots mark individual corpus callosum measurements, and the quadratic curve models the relationship between corpus callosum area and age. Standard errors are indicated by the shaded regions, indicating no detectable difference in means between typical controls and autism cases (Color figure online)
Corpus Callosum Subregions in ASD and DSM-IV Subgroups
There was no evidence for overall ASD-related differences in any Witelson corpus callosum subregions (Fig. 3, range −1.64 to 1.75 % change in area relative to controls, p values range from 0.36 to 0.99). In the DSM-IV autism subtype, there was evidence for corpus callosum area reduction in region W4 (2.4 % area reduction, p = 0.01). No strong evidence was found for other subregional changes in corpus callosum area in Autism (range −2.44 to −0.42 %, p value range 0.09–0.62), Asperger’s syndrome (range −1.44 to 2.86 %, p value range 0.36–0.92) or PDD-NOS groups (−4 to 3.9 %, p value range 0.2–0.85).
There is little evidence for corpus callosum subregional area changes in autism spectrum disorder or autism subtypes. The figure indicates percentage differences between controls and individuals with autism spectrum disorder are < 4 % for all corpus callosum subregions (note PDD-NOS region W1 difference was −13.2 % but was truncated to preserve formatting in the plot). Subregions were parcellated according the Witelson scheme (Color figure online)
Total Intracranial Volume
There was evidence for increased intracranial volume in autism spectrum disorder (1.73 % volume increase, p = 0.042, Fig. 4). When the subtypes are analyzed separately there is no clear indication for intracranial volume differences in any one subgroup (p range from 0.21 to 0.77), although the estimated magnitude of the difference appears greatest in the PDD-NOS group (1.8 % increase, p = 0.5).
Cross-sectional intracranial volume changes with age in autism spectrum disorder (red) and healthy controls (blue). The figure indicates there is subtle but statistically significant increased intracranial volume in individuals with autism spectrum disorder (p = 0.042). Note that the difference between ASD and controls (distance between red and blue lines) is greater in younger participants around age 10, and minimal around age 20 + , which is consistent with reports from previous studies (Redcay and Courchesne 2005). Grey shaded regions represent standard errors (Color figure online)
Discussion
Our analysis of corpus callosum mid-sagittal area did not find any evidence for autism spectrum disorder related differences in the ABIDE study cohort. A subtle reduction in corpus callosum area in the autism DSM-IV subcategory was found (−13.42 mm2, p = 0.045). The estimated effect size for overall corpus callosum area differences in autism spectrum disorders, calculated as Cohen’s d, was 0.016, and for the autism DSM-IV subcategory Cohen’s d = −0.02. Both of these values are considerably lower than the estimated effect size of 0.48 reported in a previous meta-analysis (Frazier and Hardan 2009). It is important to note that calculating Cohen’s d implies the presence of an effect, but our results show no strong evidence for a difference in corpus callosum area in ASD. Therefore the calculated d value is only useful for comparison with previous studies (Frazier and Hardan 2009) and does not imply that an effect exists. Our large n study does not agree with the majority of papers that report a reduction in overall corpus callosum area in autism spectrum disorders, although four previous studies found no differences in corpus callosum morphology in autism (Elia et al. 2000; Herbert et al. 2004; Rice et al. 2005; Tepest et al. 2010).
Our analysis of the ABIDE cohort identified a subtle intracranial volume increase in autism spectrum disorder (p = 0.042), which is consistent with previous studies. There is little clear evidence to suggest this difference is driven by any particular autism subcategory. It is interesting to note that our measured intracranial volume increase of 1.73 % is fairly close to the 1.58 % volume increase predicted by a model provided in a meta-analysis published by Redcay et al., based on an average age of 17.6 for our ASD participants (Redcay and Courchesne 2005).
One potential explanation for disagreement between the results of our analysis and some prior reports may be the high measured IQ of the ABIDE autism subjects relative to the autism population in general; the average IQ for ABIDE autism subjects was 105, compared with previously reported mean IQs for ASD of approximately 70 (Charman et al. 2011). There is a strong co-occurrence of ASD and intellectual disability, with some reports estimating that approximately half of individuals with ASD also have intellectual disability [IQ < 70, (Bertrand et al. 2001; Charman et al. 2011)]. Individuals with ASD and intellectual disability were not included in the ABIDE cohort. A potential hypothesis that explains the divergent findings is that reduced corpus callosum area reflects an underlying abnormality in brain development that may manifest as intellectual disability and autism. It should be noted that not all previous literature support this hypothesis; for example there are previous studies with similar IQ ranges to the ABIDE study that reported differences in corpus callosum area (Freitag et al. 2009), mean autism FSIQ = 101; Alexander et al. (2007), mean autism FSIQ = 107).
The use of geographically dispersed multiple international recruitment centers for the ABIDE database should be considered when interpreting the results of our study [and other studies utilizing the ABIDE database (Haar et al. 2014; Valk et al. 2015)]. A number of potential factors that may influence autism prevalence have been identified, any number of which could vary or co-occur between the sites in ABIDE and which may explain divergent findings in the literature (Hughes 2009). Bearing this caveat in mind, we believe that our analysis of imaging data from all sites in the ABIDE database is likely to provide more generalizable findings than smaller single site studies.
Many studies report that neuroanatomical abnormalities associated with autism spectrum disorder develop during prenatal and early postnatal life (Courchesne et al. 2011). The ABIDE dataset’s age range lies outside of this early developmental period and would therefore not detect previously reported neuroanatomical trajectory differences in younger patients with autism (Amaral et al. 2008). Furthermore, our cross-sectional analysis cannot examine differences in the trajectory of brain development of healthy individuals versus patients with autism which can be investigated in longitudinal studies; however, overall differences in corpus callosum areas have been reported in adolescence and young adulthood and the design of the ABIDE study is appropriate to investigate these differences.
An additional limitation for our study that should be noted is that the Witelson scheme is a geometric subdivision scheme; it is possible that this method for dividing the corpus callosum is suboptimal for detecting subtle anatomical differences that may be smaller than or overlap Witelson regions. Given that the corpus callosum is a structure that consists of white matter pathways between brain regions, diffusion weighted imaging and related tractographic methods may provide higher sensitivity for investigating brain changes in autism spectrum disorders.
In summary, our analysis presents little compelling evidence for reduced corpus callosum area in autism spectrum disorders, however there was evidence for increased intracranial volume. Our use of an objective measurement technique, blinded analysis, large sample size, and the multi-site design of the ABIDE study suggests that reduced corpus callosum area is not a consistent feature across autism spectrum disorders.
References
Alexander, A. L., Lee, J. E., Lazar, M., Boudos, R., DuBray, M. B., Oakes, T. R., et al. (2007). Diffusion tensor imaging of the corpus callosum in autism. Neuroimage, 34(1), 61–73.
Amaral, D. G., Schumann, C. M., & Nordahl, C. W. (2008). Neuroanatomy of autism. Trends in Neurosciences, 31(3), 137–145.
Ardekani, B. A. (2013). yuki module of the automatic registration toolbox (ART) for corpus callosum segmentation.
Bertrand, J., Mars, A., Boyle, C., Bove, F., Yeargin-Allsopp, M., & Decoufle, P. (2001). Prevalence of autism in a United States population: The Brick Township, New Jersey, investigation. Pediatrics, 108(5), 1155–1161.
Charman, T., Pickles, A., Simonoff, E., Chandler, S., Loucas, T., & Baird, G. (2011). IQ in children with autism spectrum disorders: Data from the special needs and autism project (SNAP). Psychological Medicine, 41(3), 619–627.
Courchesne, E., Campbell, K., & Solso, S. (2011). Brain growth across the life span in autism: Age-specific changes in anatomical pathology. Brain Research, 1380, 138–145.
Di Martino, A., Yan, C. G., Li, Q., Denio, E., Castellanos, F. X., Alaerts, K., et al. (2013). The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry, 19(6), 659–667.
Egaas, B., Courchesne, E., & Saitoh, O. (1995). Reduced size of corpus callosum in autism. Archives of Neurology, 52(8), 794–801.
Elia, M., Ferri, R., Musumeci, S. A., Panerai, S., Bottitta, M., & Scuderi, C. (2000). Clinical correlates of brain morphometric features of subjects with low-functioning autistic disorder. Journal of Child Neurology, 15(8), 504–508.
Frazier, T. W., & Hardan, A. Y. (2009). A meta-analysis of the corpus callosum in autism. Biological Psychiatry, 66(10), 935–941.
Freitag, C. M., Luders, E., Hulst, H. E., Narr, K. L., Thompson, P. M., Toga, A. W., et al. (2009). Total brain volume and corpus callosum size in medication-naive adolescents and young adults with autism spectrum disorder. Biological Psychiatry, 66(4), 316–319.
Haar, S., Berman, S., Behrmann, M., & Dinstein, I. (2014). Anatomical abnormalities in autism? Cerebral Cortex. doi:10.1093/cercor/bhu242
Herbert, M. R., Ziegler, D. A., Makris, N., Filipek, P. A., Kemper, T. L., Normandin, J. J., et al. (2004). Localization of white matter volume increase in autism and developmental language disorder. Annals of Neurology, 55(4), 530–540.
Hughes, J. R. (2009). Update on autism: A review of 1300 reports published in 2008. Epilepsy & Behavior, 16(4), 569–589.
Jenkinson, M., Bannister, P., Brady, M., & Smith, S. (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage, 17(2), 825–841.
Redcay, E., & Courchesne, E. (2005). When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biological Psychiatry, 58(1), 1–9.
Rice, S. A., Bigler, E. D., Cleavinger, H. B., Tate, D. F., Sayer, J., McMahon, W., et al. (2005). Macrocephaly, corpus callosum morphology, and autism. Journal of Child Neurology, 20(1), 34–41.
Robinson, A. (2010). Equivalence: Provides tests and graphics for assessing tests of equivalence. R package version 0.5.6.
Team, R. C. (2013). R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Tepest, R., Jacobi, E., Gawronski, A., Krug, B., Moller-Hartmann, W., Lehnhardt, F. G., & Vogeley, K. (2010). Corpus callosum size in adults with high-functioning autism and the relevance of gender. Psychiatry Research, 183(1), 38–43.
Valk, S. L., Di Martino, A., Milham, M. P., & Bernhardt, B. C. (2015). Multicenter mapping of structural network alterations in autism. Human Brain Mapping, 36, 2364–2373.
Witelson, S. F. (1989). Hand and sex differences in the isthmus and genu of the human corpus callosum. A postmortem morphological study. Brain, 112(Pt 3), 799–835.
Yushkevich, P. A., Piven, J., Hazlett, H. C., Smith, R. G., Ho, S., Gee, J. C., & Gerig, G. (2006). User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage, 31(3), 1116–1128.
Acknowledgments
Supported by Amazon Web Services Education in Research grants.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kucharsky Hiess, R., Alter, R., Sojoudi, S. et al. Corpus Callosum Area and Brain Volume in Autism Spectrum Disorder: Quantitative Analysis of Structural MRI from the ABIDE Database. J Autism Dev Disord 45, 3107–3114 (2015). https://doi.org/10.1007/s10803-015-2468-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10803-015-2468-8