Abstract
Diabetic nephropathy (DN) is a common diabetic complication. Studies show that mitophagy inhibition induced-ferroptosis plays a crucial role in DN progression. UHRF1 is associated with mitophagy and is highly expression in DN patients, however, the effect of UHRF1 on DN is still unclear. Thus, in this study, we aimed to investigate whether UHRF1 involves DN development by the mitophagy/ferroptosis pathway. We overexpressed UHRF1 using an adeno-associated virus 9 (AAV9) system in high-fat diet/streptozotocin-induced diabetic mice. Renal function index, pathological changes, mitophagy factors, and ferroptosis factors were detected in vivo. High-glucose cultured human renal proximal tubular (HK-2) cells were used as in vitro models to investigate the mechanism of UHRF1 in DN. We found that diabetic mice exhibited kidney damage, which was alleviated by UHRF1 overexpression. UHRF1 overexpression promoted PINK1-mediated mitophagy and inhibited the expression of thioredoxin interacting protein (TXNIP), a factor associated with mitochondrial dysfunction. Additionally, UHRF1 overexpression alleviated lipid peroxidation and free iron accumulation, and upregulated the expression of GPX4 and Slc7a11, indicating the inhibition effect of UHRF1 overexpression on ferroptosis. We further investigated the mechanism of UHRF1 in the mitophagy/ferroptosis pathway in DN. We found that UHRF1 overexpression promoted PINK1-mediated mitophagy via inhibiting TXNIP expression, thus suppressing ferroptosis. These findings confirmed that upregulation of UHRF1 expression alleviates DN, indicating that UHRF1 has a reno-protective effect against DN.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Diabetic nephropathy (DN) is one of the most common microvascular complications of diabetes. With the increasing incidence of diabetes, DN is the leading cause of chronic kidney disease and end-stage renal failure [1]. How to effectively treat DN has become an important issue. It is of great significance to investigate the pathogenesis of DN and find effective treatment.
Mitophagy is a form of autophagy that selectively degrades damaged or excess mitochondria. Mitophagy prevents excessive reactive oxygen species (ROS) accumulation and activates the mitochondrial apoptotic cascade, thus reducing kidney injury [2]. DN prominently exhibits an accumulation of dysfunctional mitochondria, indicating lowered or aberrant mitophagy. Ferroptosis is a novel form of programmed cell death, which participates in the pathogenesis of DN [3]. Mitophagy inhibition leads to excessive ROS production, whereas a large accumulation of ROS results in the formation of cellular ferroptosis [4, 5], indicating that the pathological process of ferroptosis is closely related to mitophagy. Moreover, the PINK1/Parkin pathway is a crucial mitophagy pathway. PINK1 activates Parkin activity resulting in ubiquitination of multiple proteins and thus promotes mitophagy [6]. Previous research has shown that PINK1/Parkin-mediated mitophagy protects renal tubular epithelial cells from ferroptosis [7], suggesting the regulation of the mitophagy/ferroptosis pathway may be a potential treatment against DN.
Ubiquitin like with PHD and ring finger domains 1 (UHRF1), an epigenetic regulator, is an important link between DNA methylation and histone modification [8]. UHRF1 knockdown activates the TGF-β signaling pathway and promotes the expression of collagen type I alpha 1 chain 1 (COL1A1, a main component of the extracellular matrix) [9]. The accumulation of extracellular matrix is also a pathological feature of DN [10]. Thioredoxin interacting protein (TXNIP) inhibits mitophagy in diabetes [11], coincidentally, UHRF1 also inhibits the expression of TXNIP by epigenetic regulation [12, 13]. Moreover, UHRF1 knockdown increases the level of oxidative stress [14, 15], and the inhibition of UHRF1 expression also aggravates the occurrence of ferroptosis [16]. Importantly, another study reports that UHRF1 is highly expressed in DN patients [17]. These findings suggest that UHRF1 may play a protective function on DN. Given the above, we speculated that UHRF1 may participate in DN progression via regulating mitophagy.
In this study, the UHRF1 overexpression mouse model of diabetes was used to investigate the role of UHRF1 in DN. Furthermore, we also determined the underlying mechanism of UHRF1 using a tubular epithelial cell model.
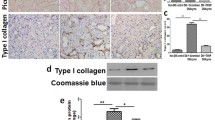
METHODS
Tissue Samples
Normal human kidney tissue samples were obtained from the nondiabetic patients with radical nephrectomy for renal carcinoma and renal biopsy specimens were collected from DN patients. Signed informed consent forms were acquired from all patients who participated. This experiment was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University and complied with the Declaration of Helsinki.
Diabetic Nephropathy Animal Models
The animal experiments were cared for according to NIH guidelines and were approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University. Seven-week-old male C57BL/6 J mice were randomized into two groups: Control and HFD-STZ groups. HFD-STZ mice were fed a high-fat diet (HFD, 60% fat) for 4 weeks. Then, they were treated with streptozotocin (STZ, 50 mg/kg/daily intraperitoneal injection) for 5 consecutive days. After STZ injection, HFD-STZ mice were fed with HFD. The fasting blood glucose (FBG) level was detected 72 h after STZ injection. Based on the prior literature [18, 19], FBG ≥ 11.1 mmol/L was considered a successful establishment of the type-II diabetes mouse model. The control mice were fed normal feed and injected with an equal volume of vehicle solution every day for five days, then continued to be fed with normal feed. After 12 weeks of feed, serum, urine, and kidney samples were collected for analysis.
For UHRF1 overexpression, mice with successful modeling were divided into three groups: HFD-STZ, HFD-STZ+NCAAV9, and HFD-STZ+UHRF1AAV9. HFD-STZ+NCAAV9 and HFD-STZ+UHRF1AAV9. Mice were administered 100 μL adeno-associated virus-9 empty vector (NCAAV9) and UHRF1 overexpressed AAV9 (UHRF1AAV9) by renal vein, respectively. HFD-STZ mice were injected with an equal volume of vehicle solution. After injection, mice continued to be fed with HFD.
Biochemical Detection
The levels of urinary albumin with Mouse Alb (albumin) ELISA kit (FineTest, Wuhan, China) and urine neutrophil gelatinase-associated lipocalin (NGAL) with Mouse NGAL ELISA Kit (FineTest, Wuhan, China) were detected. Chemiluminescence was performed to determine urine creatinine, serum creatinine (Scr), and serum blood urea nitrogen (BUN). Urine albumin/urine creatinine (UACR) was used to detect urinary protein.
Cell Culture and Transfection
Human renal proximal tubular (HK-2) cells were purchased from iCell (Shanghai, China) and cultured in the keratinocyte-serum-free medium at 37 ℃ with 5% CO2. Cells were incubated in the high-glucose (HG, 30 mM) or normal glucose (NG, 5.5 mM) medium for 72 h. NG medium contained mannitol to balance osmotic pressure. For transfection, cells were transfected with UHRF1 overexpression plasmids (UHRF1-OE), shPINK1 plasmids (shPINK1), or TXNIP overexpression plasmids (TXNIP-OE). 48 h post-transfection, cells were cultured in HG or NG medium for 72 h for further experiments.
Lipid Peroxidation Measurement
Lipid peroxidation was detected via fluorescent probe C11-BODIPY581/591 (Maokang, Shanghai, China) staining. Cells were incubated with 2 μM C11-BODIPY581/591 for 20 min. After washing, cells were collected and resuspended for flow cytometry (NovoCyte, Agilent, USA).
Immunohistochemistry
After paraffin embedding the kidney tissues, the tissues were cut into sections 5 µm thick. Then, the sections went through antigen retrieval, and incubation with 3 H2O2. Following blocking with 1% BSA, sections were incubated with primary antibodies against UHRF1 (21402-1-AP, Proteintech, RRID: AB_10860902), GPX4 (DF6701, Affinity, RRID: AB_2838663), Slc7a11 (26864-1-AP, Proteintech, RRID: AB_2880661), and PINK1 (DF7742, Affinity, RRID: AB_2841209) overnight at 4 ℃. After washing, sections were incubated with HRP-labeled goat anti-rabbit IgG for 60 min. The stained sections were developed with DAB, followed by counterstaining with haematoxylin. Images were captured using a BX53 microscope (OLYMPUS, Japan).
Periodic Acid-Schiff (PAS) Staining
Sections were incubated with periodic acid for 10 min. After washing, sections were stained with Schiff's reagent for 15 min. Then, hematoxylin solution was used to counterstain for 2 min. Sections were dehydrated in the subsequent steps: 75% ethanol for 2 min, 85% ethanol for 2 min, 95% ethanol for 2 min, absolute ethanol for 5 min, and xylene for 10 min. Images were captured by a BX53 microscope (OLYMPUS, Japan).
Western Blot
The proteins from tissues and cells were extracted and the protein quantification was conducted using a BCA protein quantification kit (Beyotime, Shanghai). Next, proteins were split with SDS-PAGE and transferred onto PVDF membranes (Millipore, USA). After blocking with 5% BAS solution for 1 h, membranes were incubated with primary antibodies against UHRF1 (21402-1-AP, Proteintech, RRID: AB_10860902), PINK1 (DF7742, Affinity, RRID: AB_2841209), Parkin (AF0235, Affinity, RRID: AB_2833410), TXNIP (18243-1-AP, Proteintech, RRID: AB_2876873), LC3 II/I (14600-1-AP, Proteintech, RRID: AB_2137737), p62 (18420-1-AP, Proteintech, RRID: AB_10694431), COX IV (66110-1-Ig, Proteintech, RRID: AB_2881509) or β-actin (66009-1-Ig, Proteintech, RRID: AB_2687938). The next day, membranes were incubated with goat anti-rabbit IgG (SA00001-2, Proteintech, RRID: AB_2722564) or goat anti-mouse IgG (SA00001-1, Proteintech, RRID: AB_2722565). Proteins were detected with enhanced chemiluminescence (ECL) reagent and the optical density value was determined using Gel-Pro-Analyzer software.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted with TRIpure solution and reverse-transcribed into cDNA using BeyoRT II M-MLV Reverse Transcriptase (Beyotime, Shanghai). Gene expression was quantified via a Real-Time PCR machine (Exicycler 96, BIONEER, Korea). The primer sequences included the homo TXNIP-specific forward 5’-ATCAGTCAGAGGCAATCA-3’ and reverse 5’-CCAGGAACGCTAACATA-3’, homo UHRF1-specific forward 5’-CTCAACTGCTTTGCTCCCA-3’ and reverse 5’-TCTTGCCACCCTTGACATT-3’, mus UHRF1-specific forward 5’-GAGCAAGAAGAAGGCAAAG-3’ and reverse 5’-ACAGGGACACCAGGGAT-3’, and mus TXNIP-specific forward 5’-TTACCCGAGTCAAAGCCGTC-3’ and reverse 5’-CCATCTCGTTCTCACCTGCT-3’. The gene expression was calculated via the 2−ΔΔCT method.
Immunofluorescence (IF)
Immunofluorescent staining of tissues was performed on paraffin-embedded tissues. Briefly, sections were immersed in xylene for dewaxing and dewaxing and rehydrated through graded ethanols. Next, sections were blocked with 1% BSA for 15 min followed by incubation with primary antibodies: LC3 (14600-1-AP, Proteintech, RRID: AB_2137737) and COX-IV (66110-1-Ig, Proteintech, RRID: AB_2881509) at 4 ℃ overnight. Then sections were incubated with FITC-labeled goat anti-rabbit IgG and Cy3-labeled goat anti-mouse IgG, followed by DAPI (Aladdin, Shanghai, China) nuclear stain. All sections were photographed under a BX53 microscope (OLYMPUS, Japan).
Oxidative Stress Detection
The levels of malondialdehyde (MDA), glutathione (GSH), and Fe2+ using the MDA assay kit (Jiancheng, Nanjing, China), GSH assay kit (Jiancheng, Nanjing, China), and ferrous ion colorimetric assay kit (Elabscience, Wuhan, China) according to the manufacturer’s instructions.
CCK-8 Assay
Cells were seeded in 96-well plates (5 × 103 cells/well) and incubated at 37 ℃ with 5% CO2. Then, 10 μl CCK-8 solution was added into each well and incubated for 2 h. The optical density (OD) value was detected at 450 nm by a microplate reader (800TS, BioTek, USA).
Chromatin Immunoprecipitation-qRT-PCR (ChIP-qPCR)
ChIP assay was conducted using a ChIP Assay Kit (Beyotime, Shanghai, China). Briefly, UHRF1 overexpression cells were collected and incubated with 1% formaldehyde for 10 min at 37 °C to cross-linking. Chromatin was sheared via sonication to produce DNA fragments. Next, chromatin was immunoprecipitated with an anti-H3K9ac antibody or IgG. The immunoprecipitated DNA was amplified with a pair of primers for the TXNIP promoter (forward 5’-CCTCCTATTTCCGTTCCACA-3’ and reverse 5’-TCAGGCCTCATTGTGTGTGT-3’). PCR amplification was carried out using 2 × Taq PCR Master Mix (Solarbio, Beijing, China). The quantitative analysis was performed on a Real-Time PCR machine (Exicycler 96, BIONEER, Korea).
Statistical Analysis
The statistical analysis was performed with GraphPad Prism 8 (GraphPad Software, Inc.). The comparison among more than two groups was used following variance (ANOVA), and the comparison between two groups was performed with a t-test. P < 0.05 was regarded as a statistically significant difference.
RESULTS
Upregulation of UHRF1 in HFD/STZ-Induced DN Mice
Previous study reports that UHRF1 is highly expressed in DN patients [14]. We also detected the protein expression of UHRF1 in DN and normal kidney tissues and the results revealed that UHRF1 was upregulated in DN tissues (Fig. 1a). Furthermore, mice were induced to DN via HFD-STZ treatment (Fig. 1b). Renal damage was assessed with UACR, NGAL/Urine creatinine ratio, BUN, and Scr. The levels of indicators were increased in HFD-STZ mice compared with control mice (Fig. 1c–f), indicating the successful establishment of DN models. Then, the UHRF1 expression in kidney tissues was detected and the results showed that UHRF1 expression both at mRNA and protein levels was upregulated in HFD-STZ mice (Fig. 1g, h). IHC staining further confirmed the higher expression of UHRF1 in HFD-STZ mice (Fig. 1i, j). These results suggested that UHRF1 was highly expressed in DN mice.
Upregulation of UHRF1 in high-fat diet (HFD)/streptozotocin (STZ)-induced diabetic nephropathy (DN) mice. (a) Expression of UHRF1 in human kidney was detected by western blot. (b) Study design overview. Seven-week-old male C57BL/6 J mice were fed with an HFD for four weeks, followed by intraperitoneal injection of STZ (50 mg/kg) for five days. Mice were fed with an HFD for 12 weeks before sacrifice. Two groups were assessed for (c) urine albumin-to-creatinine ratio (UACR), (d) urine neutrophil gelatinase-associated lipocalin (NGAL)-to-creatinine ratio (NGAL/creatinine), (e) serum blood urea nitrogen (BUN), and (f) serum creatinine (Scr). (g) The relative mRNA expression level of UHRF1 was determined by quantitative real-time PCR (qRT-PCR). (h) The protein expression of UHRF1 was determined by western blot. (i) The protein expression of UHRF1 was determined by immunohistochemistry (IHC). Bar = 200 μm. (j) Quantification of the IHC assay was represented as a percentage of UHRF1-positive cells. Data were expressed as mean ± SD, with n = 6 per group. *P < 0.05, **P < 0.01.
Overexpression of UHRF1 Alleviates DN in HFD/STZ Mice
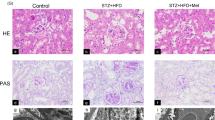
To examine the role of UHRF1 in DN, HFD-STZ mice were injected with UHRF1AAV9 for overexpressing UHRF1 (Fig. 2a). The levels of UACR, NGAL/Urine creatinine ratio, BUN and Scr were increased in HFD-STZ mice, while UHRF1 overexpression reversed the HFD-STZ-induced changes in these indicators (Fig. 2b–e). HFD-STZ mice displayed upregulated expression of UHRF1, which was further increased when UHRF1 overexpression (Fig. 2f, g). Moreover, PAS staining revealed mesangial matrix expansion and glomerular hypertrophy, early features of diabetic nephropathy, in HFD-STZ mice, whereas these injuries were ameliorated by UHRF1 overexpression (Fig. 2h). These results implied that UHRF1 overexpression alleviated DN of HFD/STZ mice.
Overexpression of UHRF1 alleviates DN in HFD/STZ mice. (a) Study design overview. Seven-week-old male C57BL/6 J mice were fed with an HFD for four weeks, followed by intraperitoneal injection of STZ (50 mg/kg) for five days. Mice were treated with adeno-associated virus-UHRF1 overexpression (UHRF1AAV9) or adeno-associated virus-negative control (NCAAV9) by left renal vein injection. Mice were maintained for 12 weeks of HFD feeding before sacrifice. Four groups were assessed for (b) UACR, (c) NGAL/creatinine), (d) BUN, and (e) Scr. (f) The protein expression of UHRF1 was determined by IHC. Bar = 200 μm. (g) Quantification of the IHC assay was represented as a percentage of UHRF1-positive cells. (h) Representative photomicrographs of periodic acid–Schiff (PAS) staining. Bar = 50 μm for tubulointerstitium, and bar = 25 μm for glomeruli. Data were expressed as mean ± SD, with n = 6 per group. *P < 0.05, **P < 0.01.
Overexpression of UHRF1 Promotes Mitophagy in DN
Mitophagy is disturbed in DN under hyperglycemic conditions [20]. PINK1/Parkin-mediated mitophagy is one of the crucial mitophagy pathways [21]. Therefore, to investigate whether UHRF1 affects mitophagy in DN, PINK1 expression was detected via IHC. Compared to the control, PINK1 expression was down-regulated after HFD-STZ treatment, which was increased when UHRF1 was overexpressed (Fig. 3a). The results of western blot further revealed that decreased expression of PINK1 and Parkin caused by HFD-STZ was reversed by UHRF1 overexpression (Fig. 3b, c). Additionally, hyperglycemia-induced TXNIP results in mitophagy inhibition in DN [11]. Our results exhibited that UHRF1 overexpression suppressed the rise of TXNIP levels caused by HFD-STZ (Fig. 3d, e). We also assessed mitophagy levels by analyzing protein levels of LC3 I/II and p62 in the mitochondria. The LC3 II was downregulated and p62 was upregulated in HFD-STZ mice, while opposite results were observed after UHRF1 overexpression (Fig. 3f, g). The results of co-localization of LC3 and COX IV further confirmed that UHRF1 overexpression rescued the inhibition of mitophagy in DN (Fig. 3h). These findings suggested that UHRF1 overexpression promoted mitophagy in DN.
Overexpression of UHRF1 promotes mitophagy in DN. (a) The protein expression of PINK1 was determined by IHC. Bar = 200 μm. (b) The protein expression of PINK1 and Parkin was determined by western blot. (c) Quantification of PINK1 and Parkin protein expressions. (d) The relative mRNA and protein levels of TXNIP were determined by qRT-PCR and western blot. (e) Quantification of TXNIP protein expression. (f) The protein expression of LC3 II/I and p62 in mitochondria was determined by western blot. (g) Quantification of LC3 II/I and p62 protein expressions. (h) Images of immunofluorescence (IF) staining of LC3 (green) and COX-IV (red) in kidney tissues. Bar = 50 μm. Data were expressed as mean ± SD, with n = 6 per group. *P < 0.05, **P < 0.01.
Overexpression of UHRF1 Inhibits Ferroptosis in DN
Ferroptosis participates in the pathogenesis of DN [3]. To evaluate the effect of UHRF1 on ferroptosis, the levels of ferrous ion (Fe2+), MDA, and GSH, three ferroptosis indicators, were detected in the renal cortex. The levels of Fe2+ and MDA were increased, and the GSH level was reduced in HFD-STZ mice. When UHRF1 was overexpressed, the levels of Fe2+ and MDA were decreased and the GSH level was increased (Fig. 4a–c). GPX4 and SLC7A11 are suppressors of ferroptosis, their protein expression in HFD-STZ mice was significantly reduced, but this trend was reversed by UHRF1 overexpression (Fig. 4d, e). Lipid peroxide is a crucial indicator of ferroptosis, which is monitored by C11-BODIPY581/591 probe. We found that the increase of lipid peroxidation caused by HFD-STZ was decreased via UHRF1 overexpression (Fig. 4f). Furthermore, the change of mitochondrial morphology is a significant feature of ferroptosis, whose main characteristic including smaller mitochondria, increased mitochondrial membrane density, and reduced mitochondrial cristae [22]. Mitochondrial damage by HFD-STZ was alleviated by UHRF1 overexpression (Fig. 4g). Collectively, these findings indicated that UHRF1 overexpression inhibited ferroptosis in DN.
Overexpression of UHRF1 inhibits ferroptosis in DN. Four groups were assessed for (a) ferrous ion (Fe2+) content, (b) malondialdehyde (MDA), and (c) glutathione (GSH). The protein expression of (d) Slc7a11 and (e) GPX4 was determined by IHC. Bar = 200 μm. (f) Lipid ROS was determined by staining with ferroptosis-dependent C11-BODIPY. (g) Mitochondrial morphology was assessed with transmission electron microscopy (TEM). Bar = 1 μm. The blue arrows point to mitochondria. Data are expressed as mean ± SD, with n = 6 per group. *P < 0.05, **P < 0.01.
UHRF1 Reverses HG-Induced Mitophagy Inhibition and Ferroptosis
To further investigate the function of UHRF1 in DN, DN cell models were constructed using HG-stimulated HK-2 cells. As shown in Fig. 5a, b, HG downregulated UHRF1 expression. We speculated that UHRF1 was upregulated in diabetic mice due to the triggering of the body's protective mechanism. However, because of the single intracellular environment, cells have difficulty responding in the same way as the body. UHRF1 overexpression cells were constructed and the successful transfection was confirmed by western blot (Fig. 5c). Then, UHRF1 overexpression cells were incubated in HG medium, and the upregulated UHRF1 expression confirmed the successful transfection (Fig. S1). The decreased expression of PINK1 and Parkin caused by HG was upregulated by UHRF1 overexpression (Fig. 5d, e). Besides, LC3 II was downregulated, and p62 expression was upregulated in cells treated with HG, whereas UHRF1 overexpression significantly reversed this trend (Fig. 5f, g). Besides, UHRF1 overexpression reduced the expression of TXNIP induced by HG (Fig. 5h, i). Moreover, reduced cell viability was observed in HG-induced cells, which was increased by UHRF1 overexpression (Fig. 5j). The morphology of mitochondria was observed and results showed that the morphology was altered after treatment with HG, indicating mitochondrial damage. This damage was rescued by UHRF1 overexpression (Fig. 5k). Furthermore, lipid ROS generation and MDA production were enhanced in HG-treated cells, while these levels were decreased when UHRF1 was overexpressed (Fig. 5l, m). These findings suggested that UHRF1 overexpression reversed HG-induced deficient mitophagy and ferroptosis. Moreover, we also detected the effect of UHRF1 knockdown on DN. UHRF1 knockdown cells were constructed and the successful transfection was confirmed by western blot (Fig. S2a). HG medium significantly reduced the expression of PINK1 and LC3 II. UHRF1 knockdown, however, resulted in a further reduction of their expression (Fig. S2b, c). The decreased cell viability caused by HG medium was further reduced by UHRF1 knockdown (Fig. S2d). Further, UHRF1 knockdown aggravated the generation of lipid ROS (Fig. S2e). These findings suggested that UHRF1 knockdown exacerbated deficient mitophagy and ferroptosis.
Overexpression of UHRF1 reverses high glucose (HG)-induced mitophagy inhibition and ferroptosis. (a) HK-2 cells were incubated with HG (30 mM) or normal glucose (NG, 5.5 mM) medium, 72 h post-incubation, the mRNA levels of UHRF1 were determined by qRT-PCR. (b) The protein levels of UHRF1 were determined by western blot. (c) HK-2 cells were transfected with UHRF1 overexpression plasmids (UHRF1-OE) or empty plasmids (EV-1), 48 h post-transfection, the protein level of UHRF1 was determined by western blot. (d) HK-2 cells were transfected with UHRF1-OE or EV-1, 48 h post-transfection, the cells were incubated with HG (30 mM) or NG (5.5 mM) for 72 h, then, the protein expression of PINK1 and Parkin was determined by western blot. (e) Quantification of PINK1 and Parkin protein expression. (f) The protein expression of LC3 II/I and p62 in mitochondria was determined by western blot. (g) Quantification of LC3 II/I and p62 protein expression. (h) The mRNA levels of TXNIP were determined by qRT-PCR. (i) The protein levels of TXNIP were determined by western blot. (j) The cell viability was determined by a CCK-8 assay. (k) Mitochondrial morphology was assessed with TEM. Bar = 500 nm. The blue arrows point to mitochondria. (l) Lipid ROS was determined by staining with ferroptosis-dependent C11-BODIPY. (m) MDA content was evaluated. Data are expressed as mean ± SD, with n = 3 per group. *P < 0.05, **P < 0.01.
UHRF1 Protects HG-Induced HK-2 Cells from Ferroptosis via Promoting PINK1-Mediated Mitophagy
To further investigate the protection effect of UHRF1 on DN, PINK1 knockdown cells were established and the successful transfection was confirmed by western blot (Fig. 6a). Then, HK-2 cells were co-transfected with shPINK1 plasmids and UHRF1 overexpression plasmids, and were incubated in HG medium. The upregulated protein expression of UHRF1 was found in cells. The expression of PINK1 and Parkin was upregulated by UHRF1 overexpression, but this trend was reversed by PINK1 knockdown (Fig. 6b, c). Additionally, UHRF1 overexpression upregulated LC3 II and downregulated p62 levels, whereas the opposite results were observed after PINK1 knockdown (Fig. 6d). The increased cell viability in UHRF1 overexpression cells was inhibited by PINK1 knockdown (Fig. 6e). Further, PINK1 knockdown promoted lip ROS accumulation (Fig. 6f) and MDA production (Fig. 6g), and reduced the levels of GSH (Fig. 6h) and GPX4 protein expression (Fig. S3) in UHRF1 overexpression cells. These results indicated that UHRF1 alleviated HG-induced ferroptosis via promoting PINK1-mediated mitophagy.
UHRF1 protects HG-induced HK-2 cells from ferroptosis via promoting PINK1-mediated mitophagy. (a) HK-2 cells were transfected with shPINK1 plasmids, 48 h post-transfection, the protein expression of PINK1 was determined by western blot. (b) HK-2 cells were co-transfected with UHRF1-OE and shPINK1 plasmids, 48 h post-transfection, the cells were incubated with HG medium for 72 h. The protein expression of UHRF1, PINK1, and Parkin was determined by western blot. (c) Quantification of UHRF1, PINK1, and Parkin protein expression. (d) The protein expression of LC3 II/I and p62 in mitochondria was determined. (e) The cell viability was determined by a CCK-8 assay. (f) Lipid ROS was determined by staining with ferroptosis-dependent C11-BODIPY. The levels of (g) MDA and (h) GSH were detected. Data are expressed as mean ± SD, with n = 3 per group. *P < 0.05, **P < 0.01.
UHRF1 Inhibits TXNIP Expression by Promoting H3K9 Deacetylation in HG-Induced HK-2 Cells
HG induces TXNIP expression by promoting the acetylation of histone (acetylated H3K9) in the TXNIP promoter region [23]. Additionally, UHRF1 recruits HDAC1 and thereby suppresses TXNIP expression through histone deacetylation [13]. To ascertain whether UHRF1 plays a protective role in DN in this way, HK-2 cells were transfected with UHRF1 overexpression plasmids and incubated in HG medium, the acetylation level of TXNIP promoter was monitored by ChIP using the antibody against acetylated H3K9 (H3K9ac) and followed by qRT-PCR. UHRF1 overexpression cells exhibited a decrease in the levels of H3K9ac acetylation on the TXNIP promoter, compared to control cells (Fig. 7a), suggesting that UHRF1 regulated TXNIP expression by histone deacetylation under stimulation with HG. Then, TXNIP overexpression cells were constructed and the successful transfection was confirmed (Fig. 7b). Furthermore, cells were co-transfected with TXNIP overexpression plasmids and UHRF1 overexpression plasmids, after incubation in HG medium, the upregulated protein expression of UHRF1 and TXNIP was observed, suggesting the successful transfection (Fig. 7c, d). Moreover, the increased level of PINK1 induced by UHRF1 overexpression was inhibited by TXNIP overexpression (Fig. 7e). These findings suggested that UHRF1 suppressed the activity of the TXNIP promoter to promote PINK1-mediated mitophagy, thus exerting a protective effect on DN.
UHRF1 inhibits TXNIP expression by promoting H3K9 deacetylation in HG-induced HK-2 cells. (a) HK-2 cells were transfected with UHRF1-OE, 48 h post-transfection, the cells were incubated with HG medium for 72 h. The qRT-PCR analysis after ChIP assay using an antibody against H3K9. IgG served as a negative control. (b) HK-2 cells were transfected with TXNIP overexpression plasmids (TXNIP-OE), 48 h post-transfection, the protein expression of TXNIP was determined. (c) HK-2 cells were co-transfected with UHRF1-OE and TXNIP-OE, 48 h post-transfection, and the cells were incubated with HG medium for 72 h. The protein expression of UHRF1 and TXNIP was detected. (d) Quantification of UHRF1 and TXNIP protein expression. (e) The protein expression of PINK1 was determined. Data are expressed as mean ± SD, with n = 3 per group. *P < 0.05, **P < 0.01. (f) The schematic diagram shows the mechanism of the protective effect of UHRF1 on DN. UHRF1 inhibits TXNIP expression by promoting H3K9 deacetylation, thus inducing PINK1-mediated mitophagy to suppress ferroptosis.
DISCUSSION
As a common complication of diabetes, DN has become one of the leading causes of death in diabetic patients [24]. In the present study, we aimed to investigate the protective effect of UHRF1 against DN and its underlying molecular mechanisms. First, we found that UHRF1 overexpression improved diabetic renal dysfunction of diabetic mice via potently reducing the levels of UACR, NGAL/urine creatinine ratio, BUN, and Scr. Also, UHRF1 overexpression reversed HG-induced mitophagy dysfunction and ferroptosis both in mice and HK-2 cells. Mechanically, UHRF1 promoted histone H3K9 deacetylation of the TXNIP promoter region, resulting in the suppression of TXNIP expression. The downregulation of TXNIP promoted PINK1-mediated mitophagy, thereby inhibiting ferroptosis (Fig. 7f). Our results confirmed that UHRF1 had a protective function in DN and may be used as a therapeutic target to prevent or treat DN.
UHRF1 is a fundamental epigenetic regulator. Recently, the upregulation of UHRF1 has been found in DN patients [17]. To explore what role UHRF1 might play in DN, we established diabetic mice, they exhibited typical features of kidney injury via the detection of renal function indexes. We also found that UHRF1 was highly expressed in diabetic mice. Importantly, UHRF1 overexpression significantly alleviated kidney injury in diabetic mice. A plethora of evidence has proved that mitophagy dysfunction is responsible for the pathologies of DN [20]. Mitophagy is a specific form of autophagy that removes defective or damaged mitochondria [25]. A reduction in mitophagy causes damaged mitochondria accumulation and oxidative stress, which contributes to the pathogenesis of DN [26]. UHRF1 depletion leads to oxidative stress [27], and it is able to inhibit TXNIP expression by epigenetic regulation [13]. Coincidentally, TXNIP knockdown alleviates mitophagy inhibition in HG-induced HK-2 cells [11]. Another study confirms that UHRF1 promotes the deacetylation of histone H3K9 of the TXNIP promoter region to mediate, thus inhibiting the expression of TXNIP [13]. We speculated that UHRF1 also suppresses TXNIP expression in DN in this way, thus promoting mitophagy. The results of the ChIP assay confirmed our conjecture, indicating that UHRF1 promoted mitophagy via inhibiting TXNIP expression. Moreover, it is reported that TXNIP overexpression represses PINK1/Parkin-mediated mitophagy [28]. PINK1/Parkin-mediated mitophagy is a crucial mitophagy pathway [29]. PINK1 localizes on the damaged mitochondrial outer membrane, then, recruits Parkin proteins to the mitochondria for activating the downstream signaling of Parkin. As an E3 ubiquitin ligase, Parkin promotes the ubiquitination of mitochondrial membrane proteins and eliminates damaged mitochondria via the activation of mitophagy [29]. The downregulation expression of PINK1 and Parkin was observed in diabetic mice, and this trend was reversed after UHRF1 overexpression, suggesting that UHRF1 overexpression promoted PINK1-mediated mitophagy. Further, upregulated expression of PINK1 caused by UHRF1 overexpression was reduced by TXNIP knockdown, confirming that UHRF1 promoted PINK1-mediated mitophagy by inhibiting TXNIP expression.
Ferroptosis is a novel form of programmed cell death, which participates in the pathogenesis of DN [3]. The pathological process of ferroptosis is closely related to mitophagy. Mitophagy inhibition causes excessive ROS production, whereas ROS accumulation eventually leads to ferroptosis [5, 30]. Further, PINK1-mediated mitophagy alleviates ferroptosis [7, 31]. Therefore, UHRF1 may inhibit ferroptosis by PINK1-mediated mitophagy. In our study, the occurrence of ferroptosis was assessed by the levels of MDA and GSH. The decreased ferroptosis was observed in diabetic mice after UHRF1 was overexpressed. Phospholipid peroxidation damage caused by the accumulation of iron-dependent ROS and lipid peroxide accumulation is a major cause of ferroptosis [32]. As a main mediator of ferroptosis, iron metabolism is critical for renal function [33]. The restriction of iron has been confirmed to delay DN development in diabetic rats [34]. Consistent with previous research, we found that UHRF1 overexpression reduced Fe2+ accumulation in kidney tissues. In addition, GPX4 normally eliminates the products of iron-dependent lipid peroxidation, which inactivation results in ferroptosis due to the accumulation of lipid ROS [35]. The lip ROS accumulation and downregulated levels of GPX4 occurred in diabetic mice, whereas UHRF1 overexpression upregulated GPX4 expression and reduced lipid ROS accumulation. Another ferroptosis inhibitor, SLC7A11, is upregulated caused by UHRF1 overexpression. Moreover, ferroptosis results in morphological changes in mitochondrial structure, which could help to further judge whether ferroptosis occurs [32, 36]. We found that the normal morphology of mitochondria was restored to some extent by UHRF1 overexpression in HG-treatment cells. The above evidence suggested that UHRF1 overexpression had an inhibition effect on ferroptosis. Consistent with previous studies, our results revealed that PINK1 knockdown reversed the inhibition effect of UHRF1 overexpression on ferroptosis, indicating that UHRF1 inhibited ferroptosis via PINK1-mediated mitophagy. Moreover, our limitation is that the effect of UHRF1 will be further illustrated if we do UHRF1 knockdown experiments in vivo, and it will be further explored in future experiments.
CONCLUSION
In conclusion, our study confirmed that UHRF1 has a protective effect on DN, possibly via inhibiting TXNIP expression, thus inducing PINK1-mediated mitophagy to inhibit ferroptosis. This work may provide a novel approach to the treatment of DN.
DATA AVAILABILITY
The data in this study is available from the corresponding author upon reasonable request.
References
Srivastava, S.P., H. Zhou, O. Setia, B. Liu, K. Kanasaki, et al. 2021. Loss of endothelial glucocorticoid receptor accelerates diabetic nephropathy. Nature Communications 12 (1): 2368. https://doi.org/10.1038/s41467-021-22617-y.
Tang, C., M.J. Livingston, Z. Liu, and Z. Dong. 2020. Autophagy in kidney homeostasis and disease. Nature Reviews Nephrology 16 (9): 489–508. https://doi.org/10.1038/s41581-020-0309-2.
Wang, Y., R. Bi, F. Quan, Q. Cao, Y. Lin, et al. 2020. Ferroptosis involves in renal tubular cell death in diabetic nephropathy. European Journal of Pharmacology 888: 173574. https://doi.org/10.1016/j.ejphar.2020.173574.
Yao, W., H. Liao, M. Pang, L. Pan, Y. Guan, et al. 2022. Inhibition of the NADPH oxidase pathway reduces ferroptosis during septic renal injury in diabetic mice. Oxidative Medicine and Cellular Longevity 2022: 1193734. https://doi.org/10.1155/2022/1193734.
Yang, Y., T. Li, Z. Li, N. Liu, Y. Yan, et al. 2020. Role of mitophagy in cardiovascular disease. Aging and Disease 11 (2): 419–37. https://doi.org/10.14336/ad.2019.0518.
Narendra, D., J.E. Walker, and R. Youle. 2012. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspectives in Biology 4 (11). https://doi.org/10.1101/cshperspect.a011338.
Lin, Q., S. Li, H. Jin, H. Cai, X. Zhu, et al. 2023. Mitophagy alleviates cisplatin-induced renal tubular epithelial cell ferroptosis through ROS/HO-1/GPX4 axis. International Journal of Biological Sciences 19 (4): 1192–1210. https://doi.org/10.7150/ijbs.80775.
Cui, Y., X. Chen, J. Zhang, X. Sun, H. Liu, et al. 2016. Uhrf1 Controls iNKT Cell Survival and Differentiation through the Akt-mTOR Axis. Cell Reports 15 (2): 256–263. https://doi.org/10.1016/j.celrep.2016.03.016.
Elia, L., P. Kunderfranco, P. Carullo, M. Vacchiano, F.M. Farina, et al. 2018. UHRF1 epigenetically orchestrates smooth muscle cell plasticity in arterial disease. The Journal of Clinical Investigation 128 (6): 2473–2486. https://doi.org/10.1172/jci96121.
Hsu, Y.H., H.H. Li, J.M. Sung, W.Y. Chen, Y.C. Hou, et al. 2017. Interleukin-20 targets podocytes and is upregulated in experimental murine diabetic nephropathy. Experimental & Molecular Medicine 49 (3): e310. https://doi.org/10.1038/emm.2016.169.
Huang, C., Y. Zhang, D.J. Kelly, C.Y. Tan, A. Gill, et al. 2016. Thioredoxin interacting protein (TXNIP) regulates tubular autophagy and mitophagy in diabetic nephropathy through the mTOR signaling pathway. Science and Reports 6: 29196. https://doi.org/10.1038/srep29196.
Kim, M. J., H.J. Lee, M.Y. Choi, S.S. Kang, Y.S. Kim, et al. 2021. UHRF1 induces methylation of the TXNIP promoter and down-regulates gene expression in cervical cancer. Molecules and Cells 44 (3): 146–59. https://doi.org/10.14348/molcells.2021.0001.
Jiao, D., Y. Huan, J. Zheng, M. Wei, G. Zheng, et al. 2019. UHRF1 promotes renal cell carcinoma progression through epigenetic regulation of TXNIP. Oncogene 38 (28): 5686–5699. https://doi.org/10.1038/s41388-019-0822-6.
Guo, Z., X. Wan, Y. Luo, F. Liang, S. Jiang, et al. 2023. The vicious circle of UHRF1 down-regulation and KEAP1/NRF2/HO-1 pathway impairment promotes oxidative stress-induced endothelial cell apoptosis in diabetes. Diabetic Medicine 40 (4): e15026. https://doi.org/10.1111/dme.15026.
Kim, J.K., G. Kan, Y. Mao, Z. Wu, X. Tan, et al. 2020. UHRF1 downmodulation enhances antitumor effects of histone deacetylase inhibitors in retinoblastoma by augmenting oxidative stress-mediated apoptosis. Molecular Oncology 14 (2): 329–346. https://doi.org/10.1002/1878-0261.12607.
Yang, R., Y. Zhou, T. Zhang, S. Wang, J. Wang, et al. 2023. The transcription factor HBP1 promotes ferroptosis in tumor cells by regulating the UHRF1-CDO1 axis. PLoS Biology 21 (7): e3001862. https://doi.org/10.1371/journal.pbio.3001862.
Peng, R., H. Liu, H. Peng, J. Zhou, H. Zha, et al. 2015. Promoter hypermethylation of let-7a-3 is relevant to its down-expression in diabetic nephropathy by targeting UHRF1. Gene 570 (1): 57–63. https://doi.org/10.1016/j.gene.2015.05.073.
Chen, Z., C. Wang, Y. Pan, X. Gao, and H. Chen. 2018. Hypoglycemic and hypolipidemic effects of anthocyanins extract from black soybean seed coat in high fat diet and streptozotocin-induced diabetic mice. Food & Function 9 (1): 426–439. https://doi.org/10.1039/c7fo00983f.
Lin, G., X. Liu, X. Yan, D. Liu, C. Yang, et al. 2018. Role of green macroalgae Enteromorpha prolifera polyphenols in the modulation of gene expression and intestinal microflora profiles in type 2 diabetic mice. International Journal of Molecular Sciences 20 (1). https://doi.org/10.3390/ijms20010025.
Kumar, A., and R. Mittal. 2018. Mapping Txnip: Key connexions in progression of diabetic nephropathy. Pharmacological Reports 70 (3): 614–622. https://doi.org/10.1016/j.pharep.2017.12.008.
Mao, Y., J. Du, X. Chen, Mamun A. Al, L. Cao, et al. 2022. Maltol Promotes Mitophagy and Inhibits Oxidative Stress via the Nrf2/PINK1/Parkin Pathway after Spinal Cord Injury. Oxidative Medicine and Cellular Longevity 2022: 1337630. https://doi.org/10.1155/2022/1337630.
Tang, D., X. Chen, R. Kang, and G. Kroemer. 2021. Ferroptosis: Molecular mechanisms and health implications. Cell Research 31 (2): 107–125. https://doi.org/10.1038/s41422-020-00441-1.
De Marinis, Y., M. Cai, P. Bompada, D. Atac, O. Kotova, et al. 2016. Epigenetic regulation of the thioredoxin-interacting protein (TXNIP) gene by hyperglycemia in kidney. Kidney International 89 (2): 342–353. https://doi.org/10.1016/j.kint.2015.12.018.
Lu, C.C., Z.B. Hu, R. Wang, Z.H. Hong, J. Lu, et al. 2020. Gut microbiota dysbiosis-induced activation of the intrarenal renin-angiotensin system is involved in kidney injuries in rat diabetic nephropathy. Acta Pharmacologica Sinica 41 (8): 1111–1118. https://doi.org/10.1038/s41401-019-0326-5.
Wang, Y., Cai J., Tang C., Dong Z. 2020. Mitophagy in acute kidney injury and kidney repair. Cells 9(2). https://doi.org/10.3390/cells9020338.
Saxena, S., A. Mathur, and P. Kakkar. 2019. Critical role of mitochondrial dysfunction and impaired mitophagy in diabetic nephropathy. Journal of Cellular Physiology 234 (11): 19223–19236. https://doi.org/10.1002/jcp.28712.
Abu-Alainin, W., T. Gana, T. Liloglou, A. Olayanju, L.N. Barrera, et al. 2016. UHRF1 regulation of the Keap1-Nrf2 pathway in pancreatic cancer contributes to oncogenesis. The Journal of Pathology 238 (3): 423–433. https://doi.org/10.1002/path.4665.
Su, C.J., Z. Shen, R.X. Cui, Y. Huang, D.L. Xu, et al. 2020. Thioredoxin-Interacting Protein (TXNIP) Regulates Parkin/PINK1-mediated Mitophagy in Dopaminergic Neurons Under High-glucose Conditions: Implications for Molecular Links Between Parkinson’s Disease and Diabetes. Neuroscience Bulletin 36 (4): 346–358. https://doi.org/10.1007/s12264-019-00459-5.
Ivankovic, D., K.Y. Chau, A.H. Schapira, and M.E. Gegg. 2016. Mitochondrial and lysosomal biogenesis are activated following PINK1/parkin-mediated mitophagy. Journal of Neurochemistry 136 (2): 388–402. https://doi.org/10.1111/jnc.13412.
Nakamura, T., I. Naguro, and H. Ichijo. 2019. Iron homeostasis and iron-regulated ROS in cell death, senescence and human diseases. Biochimica et Biophysica Acta - General Subjects 1863 (9): 1398–1409. https://doi.org/10.1016/j.bbagen.2019.06.010.
Du, Y., R. Zhang, G. Zhang, H. Wu, S. Zhan, et al. 2022. Downregulation of ELAVL1 attenuates ferroptosis-induced neuronal impairment in rats with cerebral ischemia/reperfusion via reducing DNMT3B-dependent PINK1 methylation. Metabolic Brain Disease 37 (8): 2763–2775. https://doi.org/10.1007/s11011-022-01080-8.
Dixon, S.J., K.M. Lemberg, M.R. Lamprecht, R. Skouta, E.M. Zaitsev, et al. 2012. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149 (5): 1060–1072. https://doi.org/10.1016/j.cell.2012.03.042.
Lei, P., T. Bai, and Y. Sun. 2019. Mechanisms of ferroptosis and relations with regulated cell death: a review. Frontiers in Physiology 10: 139. https://doi.org/10.3389/fphys.2019.00139.
Matsumoto, M., N. Sasaki, T. Tsujino, H. Akahori, Y. Naito, et al. 2013. Iron restriction prevents diabetic nephropathy in Otsuka Long-Evans Tokushima fatty rat. Renal Failure 35 (8): 1156–1162. https://doi.org/10.3109/0886022x.2013.819729.
Feng, H., and B.R. Stockwell. 2018. Unsolved mysteries: How does lipid peroxidation cause ferroptosis? PLoS Biology 16 (5): e2006203. https://doi.org/10.1371/journal.pbio.2006203.
Battaglia, A. M., R. Chirillo, I. Aversa, A. Sacco, F. Costanzo, et al. 2020. Ferroptosis and cancer: mitochondria meet the "iron maiden" cell death. Cells 9 (6). https://doi.org/10.3390/cells9061505.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 81570746 to G. Qin), the Medical Science and Technology Cooperation Project of Henan Province (Grant No. 2018020019 to H. Ji), the Medical Science and Technology Project of Henan Province (Grant No. 212102310196 to H. Ji).
Author information
Authors and Affiliations
Contributions
H. F. J. and G. J. Q. conceived the presented ideas. H. F. J., Y. Y. Z., X. J. M. and L. N. W. performed experiments. H. F. J., F. G., F. J. H., Y. S. and J. W. analyzed the data. H. F. J. and G. J. Q. drafted the paper. All authors have read and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ji, H., Zhao, Y., Ma, X. et al. Upregulation of UHRF1 Promotes PINK1-mediated Mitophagy to Alleviates Ferroptosis in Diabetic Nephropathy. Inflammation 47, 718–732 (2024). https://doi.org/10.1007/s10753-023-01940-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-023-01940-0