Abstract
DMP, a plant-specific membrane protein, plays a role in plant reproductive development and senescence processes. However, there is a lack of reported research on the distribution and function of the DMP gene family in sugar beet. In this study, bioinformatics methods were utilized to identify nine BvDMP family genes that were found on four chromosomes of the genome. The physicochemical properties, phylogeny, subcellular localization, gene structure, promoter regions, and replication events of these nine family genes were analyzed. RT‒qPCR was utilized to analyze the expression patterns of the nine genes within the DMP family across different tissues of sugar beet, as well as their responses to various abiotic stresses. The phylogenetic analysis indicated that all the BvDMP clusters could be categorized into six branches. BvDMPs were found to contain diverse cis-acting elements that play a role in plant responses to abiotic stresses and various phytohormones. Furthermore, expression analysis highlighted BvDMP9 as the most highly upregulated gene in reproductive organs among all members of the sugar beet DMP gene family. This finding suggests the potential involvement of BvDMP9 in the reproductive processes of sugar beet, there by providing a basis for further exploration of the functions and mechanisms of this gene family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugar beet is an important sugar crop, and approximately 20% of the global sugar production is derived from sugar beet (Stevanato P et al. 2019). However, as a biennial crop, it takes a long time to produce homozygous parents using conventional breeding methods, and the breeding process is slow. Sugar beet plants are subjected to a variety of biotic and abiotic stresses, such as drought, salinity, pests and diseases, during growth and development, which seriously affects the yield and sugar content of the crop; in contrast, haploids can be used to obtain DH (double haploid) pure lines in a single generation through chromosome doubling, which shortens the breeding time, and is a cost-effective method to breed resistant or haploid sugar beet cultivators (Zhuzhzhalova et al. 2020; Pattanayak et al. 2023). Relevant research confirmed that the DMP gene in dicotyledonous plants can induce the production of haploid plants; however, research on the DMP gene in sugar beet has not been reported.
Unknown functional structural domain families (DUFs) are protein families that contain domains of unknown function and constitute 24% of the Pfam database (Lv et al. 2023). Research has demonstrated the multifunctionality of DUF proteins. For instance, the Arabidopsis thaliana DUF584 family gene AtS40.4 is involved in seed germination and seedling growth (Shi et al. 2021). Overexpression of the A. thaliana and Brassica napus DUF2775 family gene AtDUF4 impacts plant organ size (Chen et al. 2018). Additionally, the Oryza sativa DUF726 family gene OsLDDT1 and A. thaliana DUF647 family gene RUS4 play roles in plant pollen development (Chen et al. 2020; Sun et al. 2023). Overexpression of OsSGL or heterologous expression of OsSGL in O. sativa and A. thaliana DUF1645 family genes enhances drought tolerance in transgenic plants (Cui et al. 2016). The Triticum aestivuml DUF26 family gene TaCRK3 inhibits cereal yeast mycelial growth (Guo et al. 2021). Furthermore, the O. sativa DUF668 family gene has been implicated in plant defense against pests and diseases (Zhong et al. 2019a).
DUF679 membrane proteins (DMPs), which are specific membrane proteins found in higher plants, are crucial for haploidy induction and plant growth and senescence regulation(Cyprys et al. 2019; Kasaras et al. 2010). The AtDMP protein family comprises 10 members, all with four transmembrane structural domains. AtDMP1–AtDMP7 are associated with programmed cell death, including plant senescence and stress responses in various tissues during later developmental stages (Van et al. 2006). On the other hand, AtDMP8-AtDMP10 functions in floral organs, with AtDMP8 and AtDMP9 facilitating gamete recognition, attachment, and membrane fusion during double fertilization(Takahashi et al. 2018), a process directly regulated by gamete surface proteins(Dresselhaus et al. 2016). A. thaliana HAP2/GCS1 interacts with two sperm DUF679 membrane proteins (DMP8 and DMP9) to regulate mutual recognition between sperm and oocytes (EC1) to ensure successful fertilization (Wang et al. 2022c).
The main application of DMP in breeding is haploid induction; for example, the haploid-inducing gene ZmDMP was identified on the functional locus qhir8 gene through the localization of the main effect QTL in maize, and its progeny mutants can induce the generation of maternal haploids by genetic engineering (Zhong et al. 2019b). In addition, the ZmDMP sequence is conserved between monocots and true dicots, and to verify its function in dicots, the ZmDMP homolog gene has been knocked out in A. thaliana (Zhong et al. 2020), successfully producing A. thaliana haploids. By editing the DMP homologous gene, haploid induction has been achieved in B. napus (Zhong et al. 2022b; Li et al. 2022), Nicotiana tabacum (Zhang et al. 2022), Medicago truncatula (Wang et al. 2022b) Solanum lycopersicum (Zhong et al. 2022a) and Glycine max (Nawade et al. 2023).
In this study, based on the genome sequence information of sugar beet, we investigated BvDMP genes family members and analyzed their chromosomal location, conserved structural domains, phylogeny and expression in different tissues (flower buds, stems, leaves and seeds) and under different abiotic (salt, drought and low temperature) stresses.We also investigated potential target genes for haploid breeding in sugar beet. Our findings provide a reference for further research on the function of BvDMP genes and for the haploidy breeding of sugar beet.
Materials and methods
BvDMPs identification and sequence analysis
To date, investigations of DMP genes in Zea mays (Zhong et al. 2019b), B. napus (Zhong et al. 2022b), S. lycopersicum (Zhong et al. 2022a), G. max (Nawade et al. 2023) and Citrullus lanatus (Tian et al. 2023) have been reported in the literature. To determine the sequence characteristics and related functions of sugar beet BvDMP genes, we sourced genome sequences from several databases. We obtained the genome sequences sequences of B. vulgaris and N. tabacum from the NCBI database, additionally, sequences of Z.mays (RefGen_V4), G. max(Wm82.a2.v1) and S. lycopersicum(ITAG4.0) were obtained from the Phytozome database (https://phytozome-next.jgi.doe.gov/). Genome sequences of C. lantus(Cla97_v1) and B. napus(AST_PRJEB5043_v1) were sourced from the Ensembl Plants database (http://plants.ensembl.org/index.html). The protein sequence of A. thaliana AtDMPs (TAIR, version 11, http://www.arabidopsis.org) was used as the query sequence, and the BLASTP program (score ≥ 100, e value ≤ 1e − 10) was used to search the homologous DMP sequences of the above crops. The naming of the sugar beet DMP gene refers to the naming of the A. thaliana DMP, the DMP structural domain (PF05078) was downloaded from InterproScan (http://www.ebi.ac.uk/interpro/) (Paysan-Lafosse et al. 2023) and searched against the NCBI conserved structural domain database (https://www.ncbi.nlm.nih.gov/cdd) and SMART (http://smart.embl.de/) (Letunic et al. 2021) to confirm the structural domains, then, the DMP sequences of B. vulgaris, Z. mays, G. max, B. napus, S. lycopersicum, C. lanatus and N. tabacum. Physicochemical properties such as the molecular weight, isoelectric point, and length of amino acid sequences of sugar beet BvDMP family proteins were analyzed online using the Expasy Protparam tool. The ProtScale (https://web.ExPASy.org/protscale/) program was applied to predict the hydrophilicity of sugar beet BvDMP amino acid sequences; the number of TM domains was predicted using TMHMM-2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/); subcellular localization was predicted using the SoftBerry ProtComp 9.0 (http://www.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc); and NetPhoS 3.1 (default parameters) online software was used to predict the potential sugar beet phosphorylation sites of the BvDMP protein.
Investigating phylogenetics of BvDMPs
Using MEGA 11 software, the BvDMP protein sequences were alignment with those of A. thaliana, G. max, B. napus, Z. mays, S. lycopersicum and C. lanatus, and construct an evolutionary tree using by the neighbor-joining method and the bootstrap value was set to 1000. (Trees. 1987). Finally, Modified the evolutionary tree using the online tool EvolView (http://www.evolgenius.info
/evolview/) (Subramanian B et al. 2019).
Chromosomal localization, gene duplication and gene structure analysis
The chromosomal positions of the BvDMP gene family members were extracted from the B. vulgaris genome sequence, and chromosomal gene distribution maps were constructed using TBtools (Chen et al. 2023); theconserved amino acid sequences of the proteins were analyzed online using MEME software (http://MEME-suite.org) (Bailey et al. 1994), and the parameter settings used in this study are as follows: the maximum number of different motifs is 10, and other websites default parameters.; the intron–exon structures of BvDMP genes were visualised using the GSDS (http://gsds.gao-lab.org/) (Hu et al. 2015); Extraction of gene duplication events from BvDMP gene family members with homology analysis and plotting using TBtools;Calculate the non-synonymous mutation rate (KS), non-synonymous mutation rate (ka), and the ratio of non-synonymous mutation rate to synonymous mutation rate (ka/Ks) using the Ka/Ks calculation tool (Hurst et al. 2002; Chen et al. 2023).
Prediction of the secondary structure and 3D structures of DMP proteins
The secondary and 3D structures of sugar beet BvDMP gene proteins were predicted using the online software SOPMA (https: //npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page = /NPSA/npsa_. sopma.Html) (Geourjon et al. 1995) and SWISS-MODEL (https: //swissmodel. expasy.org/ interactive) (Waterhouse et al. 2018).
BvDMP gene promoter analysis
The first 2000 bp sequence of the start codon of the upstream region of each BvDMP sequence in sugar beet was screened by the PlantCARE (Lescot et al. 2002) tool to predict and analyze cis-acting elements associated with sugar beet growth and reproduction and to group cis-regulatory elements according to their function.
Expression analysis of BvDMP genes in different tissues and organs of sugar beet
The sugar beet cultivar or N98122 was provided by the Sugar Beet Group of the Specialty Crops Institute of the Academy of Agricultural and Animal Husbandry Sciences of the Inner Mongolia Autonomous Region. The sugar beet stems, leaves, flower buds and seeds were collected at the test site, quickly frozen in liquid nitrogen and stored at −80 °C.
Using a TaKaRa RNA Extraction Kit (Code No.9767), the total RNA of each sample was extracted and reverse transcribed into cDNA using a TaKaRa M-MLV Reverse Transcription Kit and used as a template to analyze the expression of beet actin (GenBank accession no. KF214784.1). CDS was used as the template to design the internal reference gene, and PCR was performed using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus). The reaction system (20 µL) was as follows: 10 µL of SYBR Premix Ex Taq (2X), 1 µL of cDNA, 1 µL of each of the upstream and downstream primers (final concentration 0.5 µmol/L), The reaction procedure was 95 °C for 5 min, 95 °C for 10 s, 60 °C for 30 s, and 40 cycles. The dissolution curve procedure was 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s, and the signal was detected continuously. The relative expression of the BvDMP gene was calculated by the 2-ΔΔCt method and analyzed for significant differences (Livak and Schmittgen TD 2001). The primers used are listed in Table S1.
Expression analysis of BvDMPs under different abiotic stresses in sugar beet
Sugar beet N98122 seeds were sown in 6.5*5*7 (L*W*H) cm pots filled with vermiculite and incubated for 16 h/8 h (light/dark). Using Hoagland nutrient solution to maintain cultivation until the second true leaf stage. The light intensity was (450 ± 50)) μmol m−2 s−1), sunshine for 16 h per day, with a daytime temperature set at (25 ± 1) °C, a nighttime temperature set at (20 ± 1) ℃, and a relative humidity of 60–70%. In the second true leaf stage, N98122 seedlings with consistent growth were selected for processing 6 h, 12 h, 24 h and 48 h with 300 mM NaCl and 20% PEG6000, based on the wilting response of the seedlings and for processing 24 h and 48 h at 4 °C (Long et al. 2022), respectively, and untreated seedlings were as the control. Meanwhile, the second pair of true leaves from treatment frozen in liquid nitrogen and then stored them at −80 °C for reserve. Three biological replicates were used for all treatments. The total RNA extraction, reverse transcription and PCR of each sample were the same as those described in the previous subsection. Test the statistical significance between different measurement values through analysis of variance. When p < 0.05, the difference is considered statistically significant, and different letters represent significant levels of p < 0.05.
Analysis of BvDMP Gene Family Protein InteractionsIn order to investigate the interaction of BvDMP with other interacting proteins, the STRING database (https://string-db.org/) was used to construct an interaction network of BvDMP proteins by selecting homologs from sugar beet, with a confidence threshold set at 0.40.
Results
Identification and physicochemical property analysis of BvDMP gene family members
Through BLASTP comparison and structural domain analysis, using the Pfam database to remove redundant protein sequences, nine BvDMP family members were identified, namely, BvDMP2, BvDMP3, BvDMP4-A, BvDMP4-B, BvDMP4-C, BvDMP7-A, BvDMP7-B, BvDMP9 and BvDMP10. The physicochemical properties were determined separately, and the results showed that the nine BvDMP amino acid sequences of sugar beet ranged from 180 to 248 aa in length and the molecular weights ranged from 19.44 to 27.68 kDa. Among the BvDMP proteins, BvDMP4-A had the largest molecular weight, and the BvDMP2 protein had the smallest. The instability coefficients of the BvDMPs ranged from 31.72 to 47.47, and all BvDMP proteins were hydrophobic, with a grand average of hydropathy (GRAVY) greater than 0 (Fig S1). Additionally, subcellular localization predictions indicated that these proteins were located in the plasma membrane (Table S2).
Prediction of the transmembrane regions of the of nine DMP proteins using the TMHMM Server 2.0 revealed that all eight BvDMP family proteins had four transmembrane structural domains, except for the BvDMP4-A protein, which had three transmembrane structural domains. Predictive analyses using NetPhos 3.1, an online tool for identifying potential phosphorylation sites, have shed light on the sugar beet BvDMP proteins propensity for phosphorylation-an essential modification affecting protein function. Among the nine BvDMP proteins studied, we see a tapestry of potential sites ranging from a low of 24 to a high of 42 across the proteins, suggesting varying degrees of regulatory complexity.For instance, BvDMP2 set of 18 serine sites alongside 5 threonine and 6 tyrosine sites which constitute the predominant type of phosphorylation encountered in this protein.
The profile of BvDMP3 is even richer, with 25 serine sites occupying multiple positions.This abundance indicates potential multifaceted regulatory ability, supported by 14 threonine and 3 tyrosine sites, rounding out its substantial capacity for phosphorylation.Taking a close look at the BvDMP4 variants—A, B, and C—we discover a familial resemblance but with distinct personal touches. BvDMP4-A shows a diverse array with 19 serine, 13 threonine, and 3 tyrosine sites. Variants B and C, on the other hand, share an identical make-up of 23 serine and 6 threonine sites, but diverge with 4-B having a singular tyrosine site—a subtle difference that might hint at nuanced regulatory distinctions. Switching focus to BvDMP7, where A has 23 serine sites and 11 threonine sites, while B has a distribution of 14 serine sites and 12 threonine sites, supplemented with 3 tyrosine sites. The proximity in their profiles suggests a common denominator in their phosphorylation-driven roles. Remarkably, BvDMP9 phosphorylation portrait is nearly a mirror image of BvDMP4-B and C, carrying those same 23 serine and 6 threonine markers, complemented by a lone tyrosine. Last in this series, BvDMP10 displays an equitable distribution of 15 serine, 5 threonine, and 4 tyrosine sites, suggesting a potential flexibility and breadth in its regulatory interactions.
In conclusion, the study reveals a vivid mosaic of phosphorylation potential across the BvDMP proteins, painting a picture of both commonality and specificity that underlines their possible roles in sugar beet biology. The data, while complex, offers a foundational narrative for understanding the nuanced regulation of these proteins through phosphorylation. (Fig. S2).
In addition, BvDMP-9 showed more than 67% homology with A. thaliana AtDMP9 and Z. mays ZmDMP proteins and 57.26% homology with AtDMP8, which is speculated to have similar functions and can be used as a candidate gene for haploid induction in sugar beet, whereas the remaining BvDMP members showed homology with ZmDMP, AtDMP8, and AtDMP9 below 46.99% (Table 1).
Phylogenetic tree construction and Ka/Ks analysis of the DMP gene family
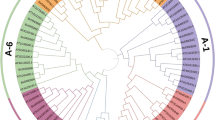
To reveal the evolutionary relationship of the beet DMP gene family, a phylogenetic tree was constructed for B. vulgaris, A. thaliana, Z. mays, G. max, B. napus, S. lycopersicum, and C. lanatus DMP protein sequences using MEGA 11 (Fig. 1). The results showed that all these DMP family genes could be classified into six subfamilies, with subfamily V having the highest number of DMP genes, containing both monocotyledonous plants (Z. mays) and dicotyledonous plants (B. vulgaris, A. thaliana, G. max, B. napus, S. lycopersicum, and C. lanatus). The sugar beet BvDMP genes were distributed across all subfamilies, among which BvDMP in subfamily I is closely related to the A. thaliana haploid-inducing genes AtDMP8 and AtDMP9 and Z. mays ZmDMP (Zm00001d044822), which are located in the same evolutionary branch and were hypothesized to be involved in haploid induction.
Analysis of conserved structural domains of proteins in the sugar beet BvDMP gene family
The conserved structural domains of the DMP protein family have been identified and analyzed in several model plants, such as A.thaliana and Z. mays. The nine BvDMP gene family members of sugar beet were analyzed by the MEME Suite motif analysis tool, and the results showed differences among the motifs of different members of the sugar beet BvDMP family. The gene structure analysis showed that all members of the family had the DUF679 structural domain (Fig. 2B) and no introns (Fig. 2D), indicating that the gene structure of each BvDMP was conserved.
Further analysis of the conserved motifs of the BvDMP proteins revealed a total of six motifs, motif 1, motif 2, motif 3, motif 4, motif 5, and motif 6 (Fig. 2A, C), of which motifs 1–4 were highly conserved among the nine sugar beet BvDMP family members, suggesting that they may be functionally similar. However, some specific motifs, such as motif 5 and motif 6 in BvDMP4-A, BvDMP4-B, and BvDMP-C (Fig. 2A), were only present in specific genes, which were hypothesized to have relevant specific functions.
Chromosomal localization analysis and colinear analysis of the BvDMP genes family
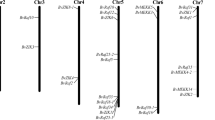
Based on the DMP gene location information in the sugar beet genome, the Tbtools tool was used to construct a schematic diagram of the DMP location on chromosomes (Fig. 3).these nine BvDMP genes were distributed across four chromosomes, with BvDMP7-B located on chromosome 1; BvDMP3 and BvDMP10 located on chromosome 4; BvDMP 9 located on chromosome 6; and BvDMP4-A, BvDMP4-B, BvDMP4-C, and BvDMP7-A located on chromosome 8. All of these BvDMP genes were located at both ends of the chromosomes.
To understand the developmental mechanisms of the sugar beet genome, Colinear genes between species were obtained by one step MCScanX analysis to explore whether gene duplication events existed between the A. thaliana and B. vulgaris.The results are shown in Fig. 4. Six homologous genes were identified in A. thaliana, among which BvDMP3, BvDMP4-A, and BvDMP9 had gene duplication events with A. thaliana AtDMP4, AtDMP5, AtDMP6, AtDMP7, AtDMP9, and AtDMP10. This indicates that these genes may have different evolutionary roles.
In the analysis of gene replication events within sugar beets, it was found that only one pair of segmental replication genes (BvDMP3 and BvDMP4-A) was present (Fig. 5). Calculation of the Ka/Ks ratio revealed that the Ka/Ks ratio was less than 1, suggesting that the gene pair underwent purifying selection during evolution.
Analysis of DMP gene covariance in Beta vulgaris. The gray line indicates the genome covariance of B. vulgaris, and the red line connecting the DMP genes indicates the duplication of DMP genes in B. vulgaris. The position of the DMP genes on the chromosomes is indicated by the short black line, and the density of the genes on each chromosome is shown at the same time
Protein structure analysis of the BvDMP gene family
Two-dimensional and three-dimensional structure prediction analyses of the proteins of the BvDMP gene family showed that the secondary structures of the proteins all contained α-helices, irregular convolutions, and extended chain irregularities. α-Helices accounted for the largest proportion of the six protein secondary structures, with a ratio of 35.02–48.79%, followed by irregular convolutions, with a ratio of 34.44–46.33%, and extended chains, with a ratio of 11.66–20.38%. of 11.66–20.38%, and β structure accounted for the lowest percentage, at 4.13–7.22% (Table S3).
Protein 3D models were constructed online using swissmodel, and the nine BvDMP genes were found to have simple and similar 3D structures (Fig. 6).
Analysis of the promoter cis-acting elements and functions of the BvDMP gene family
The exercise of a gene's function is usually regulated by its upstream promoter cis-acting elements, and analyzing cis-acting elements that may be involved in the regulation of DMP genes can help elucidate the regulatory mechanism of DMP genes and their potential functions. The 2000 bp sequence upstream of the promoter was analyzed using the online program PlantCARE. The results showed that the promoter regions of nine BvDMP gene family members contained a large number of elements responsive to phytohormones and abiotic stresses, and a total of 38 related cis-acting elements were identified, among which light-responsive elements were widely distributed in the promoter regions of all BvDMPs, such as the G-box, Box 4, ACA-motif, TCT-motif, AE-box, GATA-motif, GT1-motif, I-box, Sp1, LAMP-element, and 3-AF1 binding sites (Fig. 7).
The elements involved in abiotic stress responses are mostly cis-acting elements involved in the response to drought and low-temperature stress, such as MBS and LTR, as well as cis-regulatory elements involved in the response to the stress response elements ARE and abscisic acid and MeJA.
Involvement in plant growth and development, including involvement of the O2 site (promoter region of BvDMP2) and CAT box (promoter region of BvDMP2, BvDMP7-B, and BvDMP9), which are involved in metabolic regulation, were associated with the expression of meristematic tissues. The TGA element (promoter region of BvDMP10) and AuxRR core (promoter region of BvDMP2) were associated with the growth hormone response; the GCN4 motif (located in the promoter regions of BvDMP2, BvDMP4-B, and BvDMP10) was involved in endosperm formation; and the RY element (located in the promoter region of BvDMP10) was involved in seed-specific regulation. Among these, the GCN4 motif and RY element may be involved in the regulation of plant endosperm and seed development, suggesting that these BvDMP genes play important roles in plant reproductive development.
Analysis of the expression pattern of the BvDMP gene in different organs
To better understand the expression of BvDMP genes in different organs, we extracted RNA from the stems, leaves, flower buds and seeds of the sugar beet cultivar N98122 and assessed the expression levels of BvDMP genes in different tissues. The results showed that the expression of nine BvDMP genes in reproductive organ tissues was significantly greater than that in nutrient organs (Fig. 8). Among the BvDMP genes, BvDMP9 had the highest expression level in floral organ tissues, and the expression levels of the other eight BvDMP genes in floral organ tissues ranged from 3.51 to 11.85, which suggested that the BvDMP genes were mainly involved in the development of floral organs.
Expression analysis under different abiotic stresses
The expression patterns of BvDMPs were further analyzed under different salt, drought and low temperature stress treatments. The results showed that the expression patterns and expression levels of different BvDMP members varied greatly under different stress conditions. Under salt stress treatment conditions, BvDMP3, BvDMP7A, BvDMP7B and BvDMP9 exhibited downregulated expression with increasing treatment time; BvDMP4-B exhibited significantly greater expression than did the control at 12 h; and BvDMP4C exhibited significantly greater expression than did the control at 24 h (Fig. 9).
Under drought stress conditions, BvDMP2, BvDMP4-A, BvDMP4-B and BvDMP4-C were upregulated with increasing treatment time, while BvDMP7B and BvDMP10 were downregulated with increasing treatment time, except at some time points (Fig. 10).
Under low-temperature treatment conditions, the expression of all nine BvDMP genes in sugar beet tended to increase, and the expression of all of these genes peaked at 48 h and reached a significant difference from that of the control (Fig. 11). It was hypothesized that the different expression patterns of BvDMP genes under different stress conditions might be related to their response to adverse environmental conditions.
Prediction of the BvDMP family protein interaction network
Due to the lack of corresponding protein databases for sugar beet, in this study, we searched for homologous amino acid sequences based on the amino acid sequence of AtDMP and analyzed BvDMP protein interactions using STRING online software (https://string-db.org/). The results showed that DUO1, RALFL4, HAP2, FIM1, PEX1-2, ZAT3, GEX2, and GEX3 had interactions with the BvDMP family in the protein interaction network, which led to the speculation that sugar beet BvDMP family proteins may be the key proteins involved in flower formation and development in plants (Fig. 12).
Sublocalization analysis
Tobacco was transformed with a transient expression vector containing the BvDMP9-GFP fusion gene, and the location of the fluorescent protein was observed by confocal microscopy. The results showed that the fusion protein BvDMP9-GFP was localized to the nucleus and cell membrane (Fig. 13).
Discussion
It has been shown that haploids of maize can be induced by editing both MTL/NLD/ZmPLA1 on chromosome 1 at qhir1 and ZmDMP on chromosome 9 at qhir8 (Liu et al. 2017, 2020; Kelliher et al. 2019; Sun et al. 2022; Cheng et al. 2021; Wang et al. 2022a). The homologous ZmDMP sequences are conserved between monocotyledonous and dicotyledonous plants. The DMP homologous gene in A. thaliana was successfully knocked down in the haploids A. thaliana AtDMP8 and AtDMP9 (Wang et al. 2022a, b, c). Phylogenetic tree analysis in this study revealed that BvDMP9 of sugar beet has the highest homology with AtDMP8 and AtDMP9 of A. thaliana and belongs to subfamily IV, which is in the same subfamily as other species with haploid induction through ZmDMP homologs. In this study, we identified nine BvDMP genes from sugar beet genomic data, and the number of identified DMP genes was similar to that in A. thaliana (Zhong et al. 2020), S. lycopersicum (Zhong et al. 2022a), N. tabacum (Zhong et al. 2022b; Zhang et al. 2022), and C. lanatus (Tian et al. 2023). Analysis of the gene structure, functional domains and phylogenetic evolutionary trees of the DMP family in different species revealed that monocotyledonous and dicotyledonous plants had a common ancestor before the divergence of the DMP gene family. This family was divided into six taxa, each of which contained several subgroups, with the most members in group III and the fewest members in group VI. Moreover, group IV did not contain any sugar beet DMP family genes. During the evolutionary process, there may have been some specific gene duplications and losses in different subfamilies of the DMP family, making the number of DMP family genes different among species.
The BvDMP9 gene was analyzed using the subcellular localization method, and the BvDMP9 protein was found to be located in the cell membrane and nucleus, which was consistent with the results of homologous protein localization in maize and flax (Zhong et al. 2019b; LI et al. 2023), and the researchers in this study was hypothesized that BvDMP9 might function mainly in the plasma membrane. It was also found that there were some differences in homologous protein localization between A. thaliana and G. max. It is possible that there may be functional differences in the DMP genes of different species. It has been reported in the literature that DMP in A. thaliana has dual and time-dependent localization and is mainly expressed in membranes such as the endoplasmic reticulum and vesicular membranes (Kasaras et al. 2010), which may be related to its functions during its life cycle.
Preliminary analysis of the expression levels and expression patterns of the nine BvDMP genes in different organ tissues of sugar beet showed that they were highly expressed in the flowering organs, and previous research has shown that the parental haploid induction system was triggered in A. thaliana by stimulating the mutant DMP genes, and that such DMP proteins showed some homology in both dicotyledonous and monocotyledonous plants and that the expression patterns of the corresponding genes showed similar properties, suggesting that ZmDMP-like genes may have an important regulatory role in pollen development in Arabidopsis (Zhong et al. 2020). Another study focused on the DMP gene family in cotton and verified the expression profiles of DMP genes by RT-qPCR. The gene expression patterns assessed by qPCR were similar to the trends detected in the RNA-seq data, providing clues to the roles of DMP genes in different tissues of cotton, which also include pollen developmental regulation, and the results also showed that pollen expression reached significance compared to other tissues (Zhu et al. 2021). These suggest that they may be involved in reproductive processes and could be candidate genes for haploid induction.BvDMP displays a specific expression profile in response to stress, and in cotton (Gossypium hirsutum), it was found that DMP family genes have different expression patterns under different stress conditions such as drought, salt stress, and low temperature stress.
DMP genes may be involved in regulating osmotic pressure, ion channels, and antioxidant responses in cotton to help cotton cope with stressful environments (Zhu et al. 2021). In soybean (G. max), the study identified 14 genes in the DMP family and analysed their expression patterns at different growth stages as well as when subjected to drought and salt stress. The results showed that DMP genes in soybean are also involved in the response to abiotic stresses and that their expression increases during drought to improve drought tolerance, whereas under salt stress they may play a role by modulating ion homeostasis and stress signalling (Nawade B et al. 2023). The up-regulated expression of certain DMP genes after stress may help plants to reduce ion toxicity by enhancing the stability of cell membranes and enhancing the function of ion pumps, while others may enhance stress tolerance by regulating the synthesis of osmoprotective substances to maintain a suitable osmotic pressure in the cell. All of these changing expression patterns are an adaptive mechanism to abiotic stresses developed during plant evolution (He et al. 2024; Lv et al. 2024; Farouk et al. 2021).
Broadly speaking, the response of DMP genes to abiotic stress is usually mediated by changes in expression. In some cases, they may be upregulated to enhance certain defence mechanisms such as osmoprotection and antioxidant responses. Conversely, in other cases, the expression of certain DMP genes may be down-regulated to regulate energy utilisation and avoid cellular over-response.
It has been reported in the literature that genes without introns are more able to respond rapidly to regulate growth and developmental processes under stress (Jain et al. 2008). The absence of introns in the sequences of all nine BvDMP genes suggested that they may play an important role in adapting to abiotic stresses. Activation or repression of the downstream region of the gene promoter plays a critical role in the tissue-specific expression of components associated with plant growth and development and stress response (Liu et al. 2014). The analysis of cis-acting elements in the promoter region of DMP genes helps to elucidate BvDMP-related functions, and related research demonstrated that the light-responsive elements G-box and Box4 are essential for the regulation of light-induced transcription factors (Mallappa et al. 2006; Ezer et al. 2017; Kobayashi et al. 2012). Four BvDMP promoters have an I-box, and it has been shown that the I-box element participates in the regulation of light-induced transcription factors as a small subunit of Rubisco light regulation of photosynthesis-related genes and/or leaf-specific gene expression (Manzara et al. 1991; Shariatipour et al. 2018; Rose et al. 1999). Moreover, in various drought response investigation of drought-related elements, MBS elements were prevalent in the promoter sequences of drought-related genes (Cao et al. 2020; Avashthi et al. 2020). Most of the members of the BvDMP gene family contain the light-responsive elements G-box and Box4 and the drought-inducible cis-element MBS. In this study, RT‒qPCR analyses revealed that during drought, BvDMP4B and BvDMP4C exhibited increased expression with multiple binding sites for dehydration response elements. In addition, some stress-responsive and hormone-related elements also existed in the BvDMP gene family; for example, cis-acting elements (LTRs) involved in the low-temperature response have been identified in BvDMP7B and BvDMP9, and the relative expression of these two genes was greater than that of the other BvDMP family genes under low-temperature treatment conditions, suggesting that they were involved in the cold stress response (Wu et al. 2019; Hughes et al. 1996). Moreover, abscisic acid-responsive elements were found in the nine BvDMP genes. There is an abscisic acid response element (ABRE), which is considered an endogenous inducible regulator of abiotic stress in plants and is involved in seed dormancy and germination, stomatal closure, senescence, drought, cold and salt stress responses (Sah et al. 2016; Dar et al. 2017). Jasmonic acid response elements (CGTCA motif, TGACG motif) have high quantity of binding sites for most BvDMP genes, and jasmonic acid signaling molecules efficiently mediate defense responses to abiotic stress by inducing the expression of relevant genes (Ruan et al. 2019). In addition, stress response elements (AREs) and gibberellin response elements (P-boxes and GARE motifs) were also distributed in different numbers in each BvDMP gene, and these stress response and hormone-activated elements play important roles in developmental processes and stress resistance. Therefore, this study provides a theoretical foundation for the role of the BvDMP gene family in resistance to abiotic stress and the development of new cultivars.
Hence, this study relied heavily on bioinformatics analysis and homology comparisons. While these approaches provided valuable insights, however, experimental validation was lacking, and our next step will be to perform knockout or overexpression experiments that will solidify the hypothesized function of the BvDMP gene. This study emphasizes that the BvDMP gene will guide future research on rapid breeding and selection of resistant cultivars in sugar beet.
Conclusion
In this study, nine BvDMP genes were identified and bioinformatically assessed via genome-wide analysis, revealing their physicochemical properties and phylogenetic relationships. The nine BvDMP genes were unevenly distributed across four chromosomes of the sugar beet genome, and the physicochemical properties, phylogeny, subcellular localization, gene structure, promoter regions, and replication events of the nine gene families were analyzed in detail. Analysis of RT-qPCR results at different sites and under different stress conditions indicated that BvDMP genes play an important role in the reproductive process and in the response to abiotic stress in sugar beet. Collectively, this study led to the functional characterization of sugar beet DMP genes.This will pave the way for more future research and provide a theoretical basis for better understanding the role of BvDMP genes in stress resistance and haploid breeding of sugar beets.
Data availability
No data was used for the research described in the article.
References
Avashthi H, Pathak RK, Gaur VS et al (2020) Comparative analysis of ROS-scavenging gene families in finger millet, rice, sorghum, and foxtail millet revealed potential targets for antioxidant activity and drought tolerance improvement. Netw Model Anal Health Inform Bioinforma 9:33. https://doi.org/10.1007/s13721-020-00240-z
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Cao L, Zhang P, Lu X et al (2020) Systematic analysis of the maize OSCA genes revealing ZmOSCA family members involved in osmotic stress and ZmOSCA2.4 confers enhanced drought tolerance in transgenic arabidopsis. Int J Mol Sci 21:351. https://doi.org/10.3390/ijms21010351
Chen G, Cao X, Ma Z et al (2018) Overexpression of the nuclear protein gene AtDUF4 increases organ size in Arabidopsis thaliana and Brassica napus. J Genet Genom 45:459–462. https://doi.org/10.1016/j.jgg.2018.05.009
Chen Y-J, Yang X-X, Li W-C, Zhao S-Q (2020) Knockdown of the DUF647 family member RUS4 impairs stamen development and pollen maturation in Arabidopsis. Plant Sci 301:110645. https://doi.org/10.1016/j.plantsci.2020.110645
Chen C, Wu Y, Li J et al (2023) TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant 16:1733–1742. https://doi.org/10.1016/j.molp.2023.09.010
Cheng Z, Sun Y, Yang S et al (2021) Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol J 19:1089–1091. https://doi.org/10.1111/pbi.13584
Cui Y, Wang M, Zhou H, et al (2016) OsSGL, a Novel DUF1645 Domain-Containing Protein, Confers Enhanced Drought Tolerance in Transgenic Rice and Arabidopsis. Front Plant Sci 7. Available at: https://www.frontiersin.org/articles/https://doi.org/10.3389/fpls.2016.02001 (Accessed January 20, 2024).
Cyprys P, Lindemeier M, Sprunck S (2019) Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat Plants 5:253–257. https://doi.org/10.1038/s41477-019-0382-3
Dar NA, Amin I, Wani W et al (2017) Abscisic acid: a key regulator of abiotic stress tolerance in plants. Plant Gene 11:106–111. https://doi.org/10.1016/j.plgene.2017.07.003
Dresselhaus T, Sprunck S, Wessel GM (2016) Fertilization mechanisms in flowering plants. Curr Biol 26:R125–R139. https://doi.org/10.1016/j.cub.2015.12.032
Ezer D, Shepherd SJK, Brestovitsky A et al (2017) The G-box transcriptional regulatory code in arabidopsis. Plant Physiol 175:628–640. https://doi.org/10.1104/pp.17.01086
Farouk S, Al-Ghamdi AAM (2021) Sodium nitroprusside application enhances drought tolerance in marjoram herb by promoting chlorophyll biosynthesis and enhancing osmotic adjustment capacity. Arab J Geosci 14:430. https://doi.org/10.1007/s12517-021-06846-5
Geourjon C, Deléage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11:681–684. https://doi.org/10.1093/bioinformatics/11.6.681
Guo F, Wu T, Shen F et al (2021) The cysteine-rich receptor-like kinase TaCRK3 contributes to defense against Rhizoctonia cerealis in wheat. J Exp Bot 72:6904–6919. https://doi.org/10.1093/jxb/erab328
He S-L, Li B, Zahurancik WJ, et al (2024) Overexpression of stress granule protein TZF1 enhances salt stress tolerance by targeting ACA11 mRNA for degradation in Arabidopsis. Front Plant Sci 15. https://doi.org/10.3389/fpls.2024.1375478
Hu B, Jin J, Guo A-Y et al (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hughes MA, Dunn MA (1996) The molecular biology of plant acclimation to low temperature. J Exp Bot 47:291–305. https://doi.org/10.1093/jxb/47.3.291
Hurst LD (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 18:486–487. https://doi.org/10.1016/S0168-9525(02)02722-1
Jain M, Khurana P, Tyagi AK, Khurana JP (2008) Genome-wide analysis of intronless genes in rice and arabidopsis. Funct Integr Genom 8:69–78. https://doi.org/10.1007/s10142-007-0052-9
Kasaras A, Kunze R (2010) Expression, localisation and phylogeny of a novel family of plant-specific membrane proteins. Plant Biol 12:140–152. https://doi.org/10.1111/j.1438-8677.2010.00381.x
Kelliher T, Starr D, Su X et al (2019) One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol 37:287–292. https://doi.org/10.1038/s41587-019-0038-x
Kobayashi K, Obayashi T, Masuda T (2012) Role of the G-box element in regulation of chlorophyll biosynthesis in arabidopsis roots. Plant Signal Behav 7:922–926. https://doi.org/10.4161/psb.20760
Lescot M, Déhais P, Thijs G et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:D458–D460. https://doi.org/10.1093/nar/gkaa937
LI Wen, ZHAO Li-rong, ZHANG Jian-ping, et al. (2023) Genome-wide identification and analysis of the DMP gene family in flax (Linum usitatissimum). Acta Prataculturae Sinica 32(3):91–106. https://doi.org/10.11686/cyxb2022094
Li Y, Li D, Xiao Q et al (2022) An in planta haploid induction system in Brassica napus. J Integr Plant Biol 64:1140–1144. https://doi.org/10.1111/jipb.13270
Liu J-H, Peng T, Dai W (2014) Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol Biol Rep 32:303–317. https://doi.org/10.1007/s11105-013-0667-z
Liu C, Li X, Meng D et al (2017) A 4-bp Insertion at ZmPLA1 encoding a putative phospholipase a generates haploid induction in maize. Mol Plant 10:520–522. https://doi.org/10.1016/j.molp.2017.01.011
Liu C, Zhong Y, Qi X et al (2020) Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol J 18:316–318. https://doi.org/10.1111/pbi.13218
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Long J, Xing W, Wang Y et al (2022) Comparative proteomic analysis on chloroplast proteins provides new insights into the effects of low temperature in sugar beet. Bot Stud 63:18. https://doi.org/10.1186/s40529-022-00349-6
Lv H, Liang C, Liu W et al (2023) Multi-level biological effects of diverse alkyl chains phthalate esters on cotton seedlings (Gossypium hirsutum L.): Insights into individual, physiological-biochemical and molecular perspectives. J Hazard Mater 460:132352. https://doi.org/10.1016/j.jhazmat.2023.132352
Lv P, Wan J, Zhang C et al (2023) Unraveling the diverse roles of neglected genes containing domains of unknown function (DUFs): progress and perspective. Int J Mol Sci 24:4187. https://doi.org/10.3390/ijms24044187
Mallappa C, Yadav V, Negi P, Chattopadhyay S (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates Blue light-mediated photomorphogenic growth in arabidopsis*. J Biol Chem 281:22190–22199. https://doi.org/10.1074/jbc.M601172200
Manzara T, Carrasco P, Gruissem W (1991) Developmental and organ-specific changes in promoter DNA-protein interactions in the tomato rbcS gene family. Plant Cell 3:1305–1316. https://doi.org/10.1105/tpc.3.12.1305
Nawade B, Bosamia TC, Lee JH et al (2023) Genome-wide characterization of the soybean domain of unknown function 679 membrane protein gene family highlights their potential involvement in growth and stress response. Front Plant Sci 14:1216082. https://doi.org/10.3389/fpls.2023.1216082
Pattanayak S, Das S, Kumar S (2023) Development of stress tolerant transgenomic traits in sugar beet through biotechnological application. J Plant Protect Res 63:1–12. https://doi.org/10.24425/jppr.2023.144505
Paysan-Lafosse T, Blum M, Chuguransky S et al (2023) InterPro in 2022. Nucleic Acids Res 51:D418–D427. https://doi.org/10.1093/nar/gkac993
Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYBI is a member of a novel class of Myb-like proteins. Plant J 20:641–652. https://doi.org/10.1046/j.1365-313X.1999.00638.x
Ruan J, Zhou Y, Zhou M et al (2019) Jasmonic acid signaling pathway in plants. Int J Mol Sci 20:2479. https://doi.org/10.3390/ijms20102479
Sah SK, Reddy KR, Li J (2016) Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front Plant Sci 7. Available at: https://www.frontiersin.org/journals/plant-science/articles/10.3389/ fpls.2016.00571 (Accessed February 22, 2024).
Shariatipour N, Heidari B (2018a) Investigation of drought and salinity tolerance related genes and their regulatory mechanisms in arabidopsis. The Open Bioinformatics Journal. https://doi.org/10.2174/1875036201811010012
Shi X-P, Ren J-J, Qi H-D et al (2021) Plant-specific AtS40.4 acts as a negative regulator in abscisic acid signaling during seed germination and seedling growth in arabidopsis. Front Plant Sci 12:m622201. https://doi.org/10.3389/fpls.2021.622201
Stevanato P, Chiodi C, Broccanello C et al (2019) Sustainability of the sugar beet crop. Sugar Tech 21:703–716. https://doi.org/10.1007/s12355-019-00734-9
Subramanian B, Gao S, Lercher MJ et al (2019) Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res 47:W270–W275. https://doi.org/10.1093/nar/gkz357
Sun G, Geng S, Zhang H et al (2022) Matrilineal empowers wheat pollen with haploid induction potency by triggering postmitosis reactive oxygen species activity. New Phytol 233:2405–2414. https://doi.org/10.1111/nph.17963
Sun Z, Liu K, Chen C et al (2023) OsLDDT1, encoding a transmembrane structural DUF726 family protein, is essential for tapetum degradation and pollen formation in rice. Plant Sci 329:111596. https://doi.org/10.1016/j.plantsci.2023.111596
Takahashi T, Mori T, Ueda K et al (2018b) The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 145:dev170076. https://doi.org/10.1242/dev.170076
Tian S, Zhang J, Zhao H et al (2023) Production of double haploid watermelon via maternal haploid induction. Plant Biotechnol J 21:1308–1310. https://doi.org/10.1111/pbi.14045
Trees RP (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Van Der Graaff E, Schwacke R, Schneider A et al (2006) Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792. https://doi.org/10.1104/pp.106.079293
Wang J, Cao Y, Wang K, Liu C (2022a) Development of multiple-heading-date mtl haploid inducer lines in rice. Agriculture 12:806. https://doi.org/10.3390/agriculture12060806
Wang N, Xia X, Jiang T et al (2022b) In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol J 20:22–24. https://doi.org/10.1111/pbi.13740
Wang W, Xiong H, Cyprys P et al (2022c) DMP8 and 9 regulate HAP2/GCS1 trafficking for the timely acquisition of sperm fusion competence. Proc Natl Acad Sci USA 119:e2207608119. https://doi.org/10.1073/pnas.2207608119
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wu C, Zheng C, Ji G, Jiang P (2019) Synergistic effects of HSE and LTR elements from hsp70 gene promoter of Ulva prolifera (Ulvophyceae, Chlorophyta) upon temperature induction1. J Phycol 55:738–743. https://doi.org/10.1111/jpy.12854
Zhang X, Zhang L, Zhang J et al (2022) Haploid induction in allotetraploid tobacco using DMPs mutation. Planta 255:98. https://doi.org/10.1007/s00425-022-03877-4
Zhong H, Zhang H, Guo R et al (2019a) Characterization and functional divergence of a novel DUF668 gene family in rice based on comprehensive expression patterns. Genes 10:980. https://doi.org/10.3390/genes10120980
Zhong Y, Liu C, Qi X, Jiao Y, Wang D, Wang Y et al (2019b) Mutation of ZmDMP enhances haploid induction in maize. Nat Plants 5:575–580. https://doi.org/10.1038/s41477-019-0443-7
Zhong Y, Chen B, Li M et al (2020) A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous arabidopsis. Nat Plants 6:466–472. https://doi.org/10.1038/s41477-020-0658-7
Zhong Y, Chen B, Wang D, Zhu X, Li M, Zhang J et al (2022a) In vivo maternal haploid induction in tomato. Plant Biotechnol J 20:250–252. https://doi.org/10.1111/pbi.13755
Zhong Y, Wang Y, Chen B et al (2022b) Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum. JIPB 64:1281–1294. https://doi.org/10.1111/jipb.13244
Zhu S, Wang X, Chen W et al (2021) Cotton DMP gene family: characterization, evolution, and expression profiles during development and stress. Int J Biol Macromol 183:1257–1269. https://doi.org/10.1016/j.ijbiomac.2021.05.023
Zhuzhzhalova TP, Kolesnikova EO, Vasilchenko EN, Cherkasova NN (2020) Biotechnological methods as a tool for efficient sugar beet breeding. Vavilovskii Zh Genet 24:40–47. https://doi.org/10.18699/VJ20.593
Funding
This study was Supported by the National Key Research and Development Program of China (No.2023YFD1200703); Inner Mongolia Autonomous Region "The Open Competition Mechanism to Select the Best Candidates" project (2022JBGS0029), Inner Mongolia Autonomous Region Science and Technology Plan Project (2023KYPT0007).
Author information
Authors and Affiliations
Contributions
Pingan Han, Yue Chang, Xinrong Wu, and Xiaodong Li conducted experiments and write papers; Kuang Tang, Liang Wang, and Zhijun Xu helped analyze the data; Jing Yang, Haibo Shi, Yahui Liang, Ruifen Sun, Shaofeng Su participated in data management and contributed resources, and Ziqiang Zhang, Zengjuan Fu, Shangmin Zhao participated in bioinformatics analysis; Yuanyuan E, Wenzhe Zheng, Hui Zhang, Bizhou Zhang, and Mengyuan Sun conducted some experiments, and all authors contributed to this article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any research with human participants or animals performed by any of the authors.
Supplementary information
Supplementary Materials of Table and Figure can be found in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, P., Chang, Y., Tang, K. et al. Genome-wide identification of the sugar beet (Beta vulgaris L.) DMP gene family and its potential role in abiotic stress. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02169-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02169-y