Abstract
Wild crop relatives are unique genetic resources for crop adaptation. Increasing pressure from agriculture threatens these populations both by reducing their habitats and by creating opportunities for wild-cultivated hybridization. In this study, we assessed the diversity of 38 wild pearl millet populations covering the whole known distribution of the species in Africa, which extends from Senegal to Sudan. Using genetic analyses of 10 cultivated varieties as control, we demonstrate the high diversity harbored by these wild populations. Diversity patterns suggest a diversity hot-spot in the southern part of the wild population’s range. However, this high wild genetic diversity could partly be explained by introgression from cultivated varieties. Such introgression is widespread in the Sahel. We validate the impact of cultivated introgression on the diversity of the wild population using a genetic introgression model. The introgression distorts the real assessment of the diversity of the wild population, and the burden of this gene flow compromises the long term survival of the wild populations’ original genome. Our study also questions the long term survival of the crop’s wild relatives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the Neolithic, man has been using an increasing share of land for human mediated activities. In 2000, an estimated 50% of land surface was being used for human activities as pastureland, for cultivating crops and for cities (Goldewijk 2001; Klein Goldewijk et al. 2011), In 1700, the figure was just 10% (Goldewijk 2001; Klein Goldewijk et al. 2011).
Humans rely on a very small number of crops for their food and the diversity of these crops is less than that of their closest wild relatives. Today these wild relatives are still an important source of genetic resistance against new pathogens and could help maintain production under a changing climate (Guarino and Lobell 2011). However, the wild relatives’ habitat is threatened by the increase in agricultural land. Increasing agriculture also erodes wild diversity by providing increasing opportunities for cultivated to wild gene flow (Ellstrand et al. 1999). Some of these wild relatives’ genetic resources are now secured in ex situ collections, but ex situ collection of crop wild relative’s resources is far from complete (Castañeda-Álvarez et al. 2016). Evaluating wild relative resources in situ is urgent to assess the impact of the recent increase in land use and cultivated to wild gene flow.
Key questions concerning these wild genetic resources are (1) how does wild relatives’ population diversity vary throughout their distribution area? (2) How is this diversity reshaped by cultivated to wild gene flow? Here, we focus on wild pearl millet populations, as these are an ideal system to answer such questions because they grow in sympatric area with cultivated pearl millet but also in very isolated conditions in the Sahara. Here, we contrast areas where wild-cultivated gene flow is easy with areas where it is almost impossible. In addition, pearl millet readily outcrosses thus facilitating wild-cultivated gene flow. We sampled 48 wild populations throughout the cultivated area in the Sahel from Senegal to Sudan and from the northern limit of rainfed agriculture in West and Central Africa up to the Sahara desert. Ten cultivated varieties with a similar east–west geographical spread were included.
Materials and methods
Plant material
Ten populations of cultivated pearl millet (Pennisetum glaucum subsp. glaucum) and 38 populations of wild millet (Pennisetum glaucum subsp. monodii) collected in 10 African countries were analyzed (Figure S1). Our sampliwng area covered the central, northern and western African distribution range of pearl millet. We studied 20 individuals per population giving a total of 960 individuals. Seeds material was used for DNA extraction. Seeds are conserved in the genetic laboratory of IRD in Montpellier.
Genetic analyses
SNP analysis
We developed a set of 35 single nucleotide markers (Table S1). Single nucleotide polymorphism (SNP) was identified based on published sequence data (Clotault et al. 2012). To study wild population diversity, SNPs were designed based on their polymorphism between a set of wild individuals covering the whole range of distribution (Clotault et al. 2012). Cultivated samples were excluded to avoid an ascertainment bias to polymorphic SNPs in the cultivated samples. Genotyping was outsourced to Dr. van Haeringen Laboratorium B.V. (VHL) in Wageningen, the Netherlands.
Data analysis
Genetic diversity was estimated by determining expected heterozygosity (He), observed heterozygosity (Ho), and allelic richness (Rall, Petit and El Mousadik 1996). Expected heterozygosity is calculated by an unbiased estimator (Nei 1978):
n is the number of individuals studied in each population and Pi is the frequency of the ith allele in the same population.
Ho is the ratio of the number of heterozygous individuals observed in the population to the total number of individuals in the same population.
These parameters were compared in cultivated and wild millet groups. Computations of statistics were performed using Fstat 2.9.3 software (Goudet 2001) and statistically tested using 1000 permutations.
A Bayesian method was used to analyze genetic structure (Structure version 2.3; (Pritchard et al. 2000). We tested the admixture model with a set of populations (K) whose number ranged from one to 10. Five independent runs were performed with 50,000 burn-ins and 100,000 MCMC repetitions. The number of clusters was determined following the recommendations of Pritchard et al. (2000) and the approach developed by Evanno et al. (2005). The optimal alignment of the five independent runs for the K that was found to support the number of populations in the data was calculated using CLUMPP version 1.1.2 (Jakobsson and Rosenberg 2007). We then used Distruct (Rosenberg 2004) to display the population structure for this K. Distruct accepts additional files, thereby making it possible to control the order in which the populations are displayed, meaning populations that the Structure program assigns to the same cluster can be grouped. The same analyses were performed for K = 2 and K = 4 in order to see how the clusters progressively split.
To assess whether the variation in the genetic diversity of wild millet is affected by the ancestry of cultivated millet, we calculated a Pearson coefficient of correlation between the ancestry mean values per population (q) in the cultivated group and genetic diversity parameters (He, Ho and R) of different wild populations. The significance of the correlation was assessed using a t test. The diversity parameters of wild populations were also analyzed as a function of latitude and longitude.
Mathematical model of introgression
The consequence of introgression for diversity was modeled with a mathematical model using the STRUCTURE allele frequency estimation for K = 4. STRUCTURE estimated allele frequencies in the different “pure” populations. Based on these allele frequencies, we modeled a percentage of admixture between one of the wild population and the cultivated group (STRUCTURE assigned all cultivated populations to one group). Allelic richness and expected heterozygosity were then plotted as a function of this proportion to assess a theoretical relationship between admixture and diversity, considering the allele frequency Pij for one of the wild populations i at locus j, and considering Pcj the allele frequency of the only cultivated group at locus j. As we considered only bi-allelic loci, the frequency of one of the alleles is sufficient. The allele frequency for the j locus of a hybrid population derived from a wild population i, with an average introgression of α would be \({\text{Ph}}_{\text{ij}} = \left( {1 - \alpha } \right){\text{P}}_{\text{ij}} + \alpha {\text{P}}_{\text{cj}}\). From these allele frequencies, it is easy to deduce the expected heterozygosity He of the hybrid population for the L markers using the following formula:
The estimated number of alleles found in the hybrid population originating from wild population i considering a sample of n chromosomes, can be represented by:
The first two terms of the sum consider the probability of observing only one allele among all the chromosomes sampled, the second term the probability of observing two alleles. The sum is then averaged by the number of markers considered, L (see supplementary file).
Results
Genetic diversity
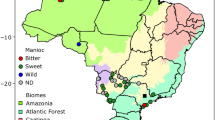
Wild populations had 50% more heterozygosity than cultivated varieties. Our final datasets contained 927 individuals (i.e. an average of 19.3 individuals per population). Allelic richness (p < 0.001), expected heterozygosity (p < 0.001) and observed heterozygosity (p < 0.003) were significantly higher in the wild millet population than in the cultivated one (Figure S2). The spatial plot of allelic richness and expected heterozygosity revealed less diversity in the center north of the Sahel (North Mali and Niger) and in the East of wild millet range (Fig. 1, Tables S2, S3). Correlation between mean ancestry and allelic richness is significant in wild population (Fig. 2). Differentiation between the 48 populations was relatively high and significant FST = 0.302 (p < 0.001). All the different markers presented a significant overall FST value, except for two with very low diversity (Table S3). Pairwise comparison of population differentiation (FST) led to 1175 significant comparisons (Bonferroni corrected p value of 4.4 10−5) and only 121 non-significant comparisons were observed (Table S4).
Genetic variation of cultivated and wild pearl millet populations from Africa. Diversity was estimated (a), using allelic richness (R) and (b) gene diversity (He). The size of the circles is proportional to the level of diversity. Blue circles represent values of diversity parameters for wild populations and red circles for cultivated populations. (Color figure online)
Genetic structure
The diversity structure of the wild populations is mainly explained by three major groups geographically structured from east to west. The population structure analyzed by STRUCTURE revealed that the likelihood ln[Pr(X/K)] values reached a plateau at K = 4 (Figure S3a), and using the Evanno et al. (2005) approach, a peak was also found at K = 4 (Figure S3b). At K = 4, all the cultivated millet populations were grouped in the same cluster whereas the wild populations were grouped in three distinct clusters (Fig. 3). The first wild cluster corresponded to samples collected in the western part of West Africa (Senegal, Mauritania and western Mali), the second from the central part of West Africa (Mali and Niger) and the last from the eastern part of the Sahel (Sudan and Chad).
Inference of populations in K clusters. Clusters are represented by colors and populations are represented by columns for (a) K = 4, (b) K = 3, (c) K = 2. The population’s ancestry was calculated as the average ancestry of all the individuals in each population. In each column, the extent of the component colors indicates the magnitude of the membership corresponding to each cluster. The name of country from which each population was sampled is labeled below the figure, and the type of each population is labeled at the top of the figure. (Color figure online)
Wild population diversity decreased from west to east, and increased with higher cultivated introgression. Diversity was higher in the western part of the area than in the eastern part (Figure S2). This result held whatever the metric (Ho, F1,34 = 15.7 p < 0.001; He F1,34=20.3 p < 0.001; Rall F1,34 = 16.5 p < 0.001) used for diversity assessment. Using K = 4 to estimate the average cultivated ancestry (Fig. 4) also explained increased diversity (Ho, F1,34 = 7.8 p < 0.01; He F1,34 = 9.2 p < 0.005; Rall F1,34 = 8.8 p < 0.006). Latitude was not correlated with variation in diversity (Table S5). The average ancestry per individual varied greatly (Fig. 4), but introgressed individuals were observed throughout the Sahel, from Senegal to Sudan. The vast majority of populations showed modest average introgression but a few populations showed relatively strong average admixture, just short of 50% between wild and cultivated populations in the western and central parts of West Africa.
Predicted introgression-diversity relationship
Modeling of wild to cultivated gene flow validated the expected relationship between cultivated gene flow and wild population diversity for moderate gene flow. We modeled expected diversity (Fig. 5) from gene flow from cultivated to wild populations. For relatively low cultivated ancestry in the wild population (< 40%) i.e. high wild ancestry (from 60 to 100%), there was a negative relationship between wild ancestry and expected heterozygosity (A) or allelic richness (B). A positive relationship was thus observed between cultivated ancestry and wild diversity. For higher cultivated ancestry (and hence low wild ancestry) diversity decreased and then tended to low cultivated diversity as wild ancestry dropped to 0%. The pattern for expected heterozygosity was similar whatever the wild group considered (western, central, eastern part of the study area), however the pattern was less clear in the groups in the western and central parts. The relationship was less smooth for allelic richness than for heterozygosity (Fig. 5). However this pattern was expected, since even low gene flow can contribute new alleles, and as a result, can rapidly increase allelic richness. The western and eastern patterns of allelic richness were quite similar. When wild ancestry reached 100%, allelic richness decreased, meaning that some specific alleles were present in the cultivated group but absent from the two specific wild groups. The pattern for the central area was different, and was almost flat when wild ancestry reached almost 100%. A flat pattern shows that most of the wild alleles are present in the cultivated sample, in other words, that the cultivated sample has no specific alleles present in the wild central group (Fig. 5).
Model of the impact of cultivated ancestry on wild populations. Calculated impact of the percentage of wild ancestry ranging from 0 (cultivated population) to 1 (wild population) on (a) expected heterozygosity, (b) allelic richness. The curve represents the ancestry in a wild group (green) in the eastern part of the African Sahel, in a group (blue) in the central part (Niger/Mali), and in a group (red) in the western part. The formula used for this estimation is described in the text. For expected heterozygosity, a higher cultivated ancestry in the wild population (i.e., a population with an ancestry ranging from 60 to 100%) led to significantly higher heterozygosity. This effect leveled off and then decreased with an increase in cultivated ancestry. Similarly, for allelic richness, low cultivated introgression generally led to an increase in allelic richness. When cultivated ancestry was very high, there was a drastic drop in allelic richness. One of the populations where the effect of ancestry was less marked is the central group (blue) both in heterozygosity and allelic richness. (Color figure online)
Discussion
In this study, we found two specific diversity patterns, a longitudinal decline from west to east and a hot-spot of diversity associated with local introgression with cultivated populations. The longitudinal pattern could be the consequence of the evolutionary history of wild pearl millet. Two hypotheses can be proposed for the origin of this pattern: first that the species originated in the western part and spread eastwards, leading to successive bottlenecks. The second hypothesis is not linked to any particular origin, but rather to the occurrence of more favorable long-term environmental conditions in the western part leading to a larger effective size. The patterns raise interesting questions about the evolutionary history of wild pearl millet, but one of the most striking results is the occurrence of widespread gene flow.
This gene flow has had a direct positive impact on the observed diversity in the wild population. Introgression of cultivated alleles into wild populations could lead to an increase in the apparent diversity of the populations concerned. Simulations easily explain this phenomenon, which required two specific patterns: one, significant differentiation between wild and cultivated populations; the other, the existence of moderate gene flow. Differentiation between wild and cultivated plants was observed here, but is a quite common phenomenon (Ellstrand et al. 1999). The other condition that needs to be met is the existence of a moderate cultivated ancestry in the wild population. If the gene flow is too high, one would expect differentiation between wild and cultivated populations to be eroded, and, given the large size of the cultivated population, the wild population to possibly be wiped out by the strong gene flow from the cultivar to the wild population. Several factors might have maintained a moderate gene flow in pearl millet: recurrent counter-selection of alleles from the cultivated population, imperfect overlapping of flowering time in cultivated and wild populations, or the existence of genetic factors leading to some form of incompatibility.
The observed patterns suggest that the differentiation continues to be maintained locally to a certain extent. We were able to establish that wild-cultivated hybridization is very common. The number of wild plants with 50% ancestry suggests F1 hybridization, and this was what was observed in different cases.
This snapshot study does not enable the evaluation of the long term consequence of gene flow between cultivated and wild pear millet. However, the relatively common occurrence of hybridization throughout the Sahel questions the long term survival of these wild populations. Our results point to the need to also study the diversity of wild relatives separately from the diversity of cultivated populations. Only the use of wild diversity coupled with cultivated varieties will make it possible to untangle regional patterns of diversity (i.e. the eastern part from the western part) and cultivated/wild gene flow in the observed diversity hot-spot of wild populations. Studying domestication without using a model that includes gene flow from wild to cultivated populations (Oumar et al. 2008; Clotault et al. 2012) could also be misleading. New modeling approaches like Approximate Bayesian computation could model gene flow from wild to cultivated populations for the inference of crop evolutionary history, which would be of particular interest. However even with a suitable model, if the wild population is partly introgressed, the signal of inference of recent gene flow or ancient origin of domestication might be partly hidden. Our result may explain the diversity of the proposed origin of cultivated pearl millet in Africa from Senegal to Sudan (Tostain 1992; Oumar et al. 2008).
Is the higher diversity found in wild populations only detrimental? Genetic diversity of populations is a metric used to assess relative population size (Ewens 2004), relative fitness (Reed and Frankham 2003) and also adaptive capacity. However, meta-analysis mainly supports its use as an estimator of relative fitness (Reed and Frankham 2003; Mittell et al. 2015). Its use as an estimator of population effective size requires the population to be isolated (Ewens 2004), which is rarely the case. Finally, meta-analysis cannot confirm a correlation of diversity and the adaptive capacity of a population (Reed and Frankham 2003; Mittell et al. 2015). However, the relationship has been demonstrated in very specific case studies (Pujol and Pannell 2008). Thus, the relationship between diversity and adaptive capacity is still the subject of debate. Here, higher diversity certainly led to higher relative fitness and might consequently allowed specific adaptation to occur. The consequence is that these populations will persist better in the future. However, introgression by cultivated varieties also means they can lose a significant proportion of their wild diversity. These populations thus present a less unique set of alleles than the cultivated population. A part of the wild allele has been lost. If gene flow continues to intensify, this relative loss could be even greater. It is thus very important to either collect more today, or to maintain an isolated specific regional reserve of wild diversity. Again, our results provide only a snapshot of wild diversity and we lack a long term perspective on the persistence of these populations.
In conclusion, we have demonstrated that cultivated to wild gene flow is possible in pearl millet in its distribution area in Africa. This gene flow could distort the evaluation of diversity in these wild populations, a measure that is often used to address their conservation status and value. As we use more and more land to cultivate crops, the opportunities for wild-cultivated gene flow are enhanced, and will have practical consequence for the survival of genetically original wild populations. The phenomenon we observed here is not certainly restricted to pearl millet, and thus calls into question the conservation status of populations of crop wild relatives at global scale.
References
Castañeda-Álvarez NP, Khoury CK, Achicanoy HA et al (2016) Global conservation priorities for crop wild relatives. Nat Plants 2:16022. https://doi.org/10.1038/nplants.2016.22
Clotault J, Thuillet A-C, Buiron M et al (2012) Evolutionary history of pearl millet (Pennisetum glaucum [L.] R. Br.) and selection on flowering genes since its domestication. Mol Biol Evol 29:1199–1212. https://doi.org/10.1093/molbev/msr287
Ellstrand NC, Prentice HC, Hancock JF (1999) Gene flow and introgression from domesticated plants into their wild relatives. Annu Rev Ecol Syst 30:539–563. https://doi.org/10.1146/annurev.ecolsys.30.1.539
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Ewens WJ (2004) Mathematical population genetics. Springer, New-York, p 417p
Goldewijk KK (2001) Estimating global land use change over the past 300 years: the HYDE Database. Glob Biogeochem Cycles 15:417–433. https://doi.org/10.1029/1999GB001232
Goldewijk KK, Beusen A, van Drecht G, de Vos M (2011) The HYDE 3.1 spatially explicit database of human-induced global land-use change over the past 12,000 years. Glob Ecol Biogeogr 20:73–86. https://doi.org/10.1111/j.1466-8238.2010.00587.x
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet (1995)
Guarino L, Lobell DB (2011) A walk on the wild side. Nat Clim Change 1:374–375. https://doi.org/10.1038/nclimate1272
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinform Oxf Engl 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Mittell EA, Nakagawa S, Hadfield JD (2015) Are molecular markers useful predictors of adaptive potential? Ecol Lett 18:772–778. https://doi.org/10.1111/ele.12454
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Oumar I, Mariac C, Pham J-L, Vigouroux Y (2008) Phylogeny and origin of pearl millet (Pennisetum glaucum [L.] R. Br.) as revealed by microsatellite loci. TAG Theor Appl Genet Theor Angew Genet 117:489–497. https://doi.org/10.1007/s00122-008-0793-4
Petit RJ, El Mousadik A (1996) High level of genetic differentiation for allelic richness among populations of the argan [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pujol B, Pannell JR (2008) Reduced responses to selection after species range expansion. Science 321:96. https://doi.org/10.1126/science.1157570
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237. https://doi.org/10.1046/j.1523-1739.2003.01236.x
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Tostain S (1992) Enzyme diversity in pearl millet (Pennisetum glaucum L.): 3. Wild millet. TAG Theor Appl Genet Theor Angew Genet 83:733–742. https://doi.org/10.1007/BF00226692
Acknowledgements
JL and YV are supported by the ARCAD project funded by the Agropolis Fondation. AA and YV are supported by the JEAI AVACLI funded by IRD.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Assoumane, A., Blay, C., Alio Sanda, AK. et al. Wild crop relative populations hot-spots of diversity are hot-spots of introgression in the case of pearl millet. Genet Resour Crop Evol 65, 1187–1194 (2018). https://doi.org/10.1007/s10722-018-0607-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-018-0607-8