Abstract
State-of-the-art production technologies for conjugate vaccines are complex, multi-step processes. An alternative approach to produce glycoconjugates is based on the bacterial N-linked protein glycosylation system first described in Campylobacter jejuni. The C. jejuni N-glycosylation system has been successfully transferred into Escherichia coli, enabling in vivo production of customized recombinant glycoproteins. However, some antigenic bacterial cell surface polysaccharides, like the Vi antigen of Salmonella enterica serovar Typhi, have not been reported to be accessible to the bacterial oligosaccharyltransferase PglB, hence hamper development of novel conjugate vaccines against typhoid fever. In this report, Vi-like polysaccharide structures that can be transferred by PglB were evaluated as typhoid vaccine components. A polysaccharide fulfilling these requirements was found in Escherichia coli serovar O121. Inactivation of the E. coli O121 O antigen cluster encoded gene wbqG resulted in expression of O polysaccharides reactive with antibodies raised against the Vi antigen. The structure of the recombinantly expressed mutant O polysaccharide was elucidated using a novel HPLC and mass spectrometry based method for purified undecaprenyl pyrophosphate (Und-PP) linked glycans, and the presence of epitopes also found in the Vi antigen was confirmed. The mutant O antigen structure was transferred to acceptor proteins using the bacterial N-glycosylation system, and immunogenicity of the resulting conjugates was evaluated in mice. The conjugate-induced antibodies reacted in an enzyme-linked immunosorbent assay with E. coli O121 LPS. One animal developed a significant rise in serum immunoglobulin anti-Vi titer upon immunization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Typhoid fever remains a serious public health problem of which there are 22–33 million cases occurring each year, including about 216,000–500,000 deaths [1]. The causative agent of this human systemic infection, S. enterica subspecies I serovar Typhi, is feco-orally transmitted through contaminated water and food. Hence, typhoid fever is endemic in less developed areas where sanitary conditions remain poor. This includes many countries of Asia, Africa and South America, where schoolchildren and young adults are most frequently affected [2]. Antimicrobial treatment of typhoid fever has become increasingly complicated through the emergence of multidrug resistant strains of S. Typhi [3].
Vaccination of high-risk populations is considered the most promising strategy for the control and prevention of typhoid fever. Currently, there are two licensed typhoid vaccines: the orally administered, live attenuated whole cell vaccine Ty21a and the purified Vi polysaccharide parenteral vaccine. The Ty21a vaccine has several disadvantages: (i) The mutations contributing to the attenuated phenotype of this S. Typhi strain are not fully defined [4], (ii) attenuated strains could theoretically revert to virulence and (iii) Ty21a is only modestly immunogenic and requires three to four initial doses and boosters every 5 years [5–9]. The usefulness of the Vi polysaccharide vaccine is limited by its age-related immunogenicity and the fact that immune responses against polysaccharides are T cell independent. Therefore, immunological memory cannot be established and revaccination does not elicit any booster response [10, 11]. Due to these drawbacks, the replacement of current typhoid vaccines with well defined, well tolerated and highly immunogenic vaccines is desirable.
The disadvantages of a polysaccharide vaccine can be overcome by conjugating the carbohydrate to a protein carrier (conjugate vaccine). Upon conjugation, the polysaccharide behaves like a T cell dependent antigen. It has been shown that purified Vi polysaccharide covalently linked to recombinant Pseudomonas aeruginosa exotoxin A (EPA) induces a protective immune response against S. Typhi in young children [12, 13].
However, production of conjugate vaccines is a complex, multi-step process. First, separate bacterial strains producing the recombinant protein carrier and the polysaccharide antigen have to be cultivated. The polysaccharide and the protein carrier have to be purified by different procedures, before the two components are chemically coupled. The last step involves additional purification steps for obtaining the final product. This laborious production process has disadvantages: (i) several purification steps are required, where considerable losses might occur and (ii) due to the random nature of chemical coupling the product is not a uniform structure but a mixture of different glycoconjugates, with potentially different efficacy profile.
A novel approach to produce conjugate vaccines is based on the bacterial N-linked glycosylation system first described in C. jejuni [14, 15]. This protein glycosylation system can be functionally transferred into E. coli to produce specified glycoproteins in vivo [15]. It has been shown that various bacterial polysaccharide structures assembled on the lipid carrier undecaprenyl pyrophosphate (Und-PP) can be transferred to selected periplasmic proteins by the oligosaccharyltransferase PglB of C. jejuni [16, 17]. Furthermore, it is possible to produce recombinant glycoproteins by grafting the consensus sequence for N-glycosylation (D/E-Z-N-X-S/T, where Z and X can be any amino acid except proline) into proteins, which are otherwise not glycosylated [18]. This cost-efficient in vivo production of glycoconjugates represents an alternative to the conventional manufacturing process.
Vi is a linear, acidic homopolymer of α1-4-linked N-acetylgalactosaminuronic acid (D-GalNAcA) residues, 60–90 % O-acetylated at C-3 (Fig. 1). Immunogenicity of the Vi is closely related to its degree of O-acetylation [19, 20]. Analysis of the bioinformatics signature of the Vi biosynthetic gene cluster (viaB) highlighted the presence of a putative ABC transporter and the absence of a wzy polymerase. Therefore, biosynthesis of Vi is thought to be similar to group 2 capsular polysaccharides (CPS). This class of CPSs is most likely not assembled on Und-PP and no periplasmic intermediates exist [21], which are both prerequisites for being a substrate of PglB. However, Und-PP linked polysaccharides exist in bacteria that resemble the Vi antigen. The O antigen of E. coli O121 (identical to Shigella dysenteriae type 7) contains an N-acetylgalactosaminuronamide (D-GalNAcAN) residue, partially O-acetylated at the C-3, which is α1-4-linked to a D-GalNAcA (Fig. 1) [22]. The O antigen cluster of E. coli O121 contains the gene wbqG, encoding a glutamine-dependent amidotransferase involved in the biosynthesis of UDP-D-GalNAcAN [23]. It has been shown that the O antigen of an E. coli O121 wbqG mutant contains D-GalNAcA (30–40 % O-acetylated at C-3) instead of D-GalNAcAN [23]. Hence, this mutant O polysaccharide contains structural similarities to the Vi (Fig. 1).
Structure of the S. Typhi Vi polysaccharide and the repeating unit of the E. coli O121 O antigen. Mutation of the O121 O antigen cluster encoded gene wbqG results in expression of a modified O polysaccharide structure. GalNAcA: 2-acetamido-2-deoxy-D-galacturonic acid; GalNAcAN: 2-acetamido-2-deoxy-D-galacturonamide; Qui4N: 4-amino-4,6-dideoxy-D-glucose
In this study the possibilities of producing a typhoid conjugate vaccine using the bacterial N-glycosylation system were exploited. The cross-reactivity of the E. coli O121 wbqG mutant O antigen with antibodies raised against the Vi is reported. Furthermore, glycoconjugates were produced and their immunogenicity was analyzed in mice.
Results
Analysis of the E. coli O121 wbqG mutant O polysaccharide
First we examined whether the O polysaccharide produced by an E. coli O121 wbqG mutant would be recognized by antibodies specific for the S. Typhi Vi capsular polysaccharide. Therefore, the E. coli O121 O antigen gene cluster was cloned, and the open reading frame of wbqG was interrupted by insertion of a STOP codon containing oligocassette. The cloned plasmids were transformed into the E. coli K-12 strain W3110 and the lipopolysaccharide (LPS) was analyzed by SDS-PAGE and staining with silver, or after transferring to a nitrocellulose membrane by Western blot (Fig. 2a). As previously reported, mutation of the wbqG gene did not abolish O antigen expression [23]. However, the LPS profile of the wbqG mutant visualized in the silver-stained polyacrylamide gel differed from the wild type in several points: (i) the staining of polymerized O antigen containing bands is fainter relative to wild type LPS, (ii) the band consisting of one O antigen repeating unit attached to the lipid A-core (core + 1RU) stained more intensely and (iii) the O antigen containing bands migrated faster than the equivalent bands of the wild type LPS. It was estimated that both LPS profiles contained an average of 12 O antigen repeat units attached to the lipid A-core by analyzing an overexposed silver-stained SDS-PAGE gel. Western blot analysis of the LPS revealed that anti-O121 sera reacted with the wbqG mutant O antigen. Interestingly, the wbqG mutant O polysaccharide was recognized by anti Vi serum.
O polysaccharide analysis of E. coli O121 and its wbqG mutant derivative. a LPS from E. coli W3110 cells expressing the O121 wild type O antigen gene cluster or its wbqG mutant derivative was separated by SDS-PAGE and stained with silver or after transfer to a nitrocellulose membrane detected with anti-O121 and anti-Vi antibodies. Mutation of wbqG results in the assembly of a modified O antigen reactive with anti-Vi sera. b Und-PP-linked glycans were extracted from E. coli SCM6 cells expressing the O121 wild type O antigen gene cluster or its wbqG mutant derivative followed by 2AB labeling and separation by normal phase HPLC using a GlycoSep N column. Individual peak fractions were analyzed by mass spectrometry and the identified glycan structures are indicated. (black square): N-acetylhexosamine; (black right-pointing triangle): dideoxyhexosamine; (white diamond): hexuronic acid; (white diamond)N: hexuronamide; Ac acetyl, NAc N-acetyl
In order to confirm the structure of the expressed O antigen repeat unit and to determine the degree of O-acetylation, glycolipids were extracted from E. coli SCM6 strains expressing either the O121 wild type or the wbqG mutant O antigen. The lipid-linked oligosaccharides were purified using a C18 SepPak column and treatment with mild acid specifically released Und-PP-linked glycans. After an additional purification step using again a C18 SepPak column, the glycans were labeled with 2-aminobenzamide (2AB) and subsequently resolved by normal phase HPLC using a GlycoSep N column. Figure 2b shows a section of the chromatogram where single repeat units and short polymerized O antigens are expected to elute. Fractions containing putative 2AB-labeled glycan species were analyzed by mass spectrometry (MS) (Fig. 3), and the glycan structures identified by MS are illustrated in Fig. 2b.
CID MS-MS spectra of glycan species separated by normal phase HPLC. The CID MS-MS spectra correspond to the glycan species identified in the individual peak fractions seen in Fig. 2b with the retention times: (a) 58.8 min, (b) 65.1 min, (c) 67.2 min, and (d) 73.5 min
The chromatogram of the 2AB-labeled glycans prepared from SCM6 cells expressing the O121 wild type O antigen, featured a peak eluting at 58.8 min. In this peak fraction a molecule with a mass-to-charge ratio (m/z) of 1083 was identified. The peak fraction with the retention time of 65.1 min contained mainly a species with m/z of 1041. This detected m/z corresponded to the single-charged sodium adduct of a 2AB-labeled, non-acetylated O121 wild type subunit. The difference between the two detected masses corresponded to 42 Da, which is the mass difference between an O-acetyl and a hydroxyl group. These two species were subjected to collisionally induced dissociation (CID) MS-MS. The series of single-charged fragment ions obtained from the precursor with m/z of 1083 (Fig. 3a) was consistent with glycosidic cleavage products from the 2AB-labeled O121 wild type O antigen repeat unit, containing an O-acetyl group at residue c. Whereas the CID MS-MS spectra of the molecular species with m/z of 1041 corresponded to the non-acetylated 2AB-labeled O121 subunit (Fig. 3b).
The chromatogram of the 2AB-labeled glycans prepared from SCM6 cells expressing the wbqG mutant polysaccharide revealed two prominent peaks. In the peak fraction with the retention time of 67.2 min a molecule with m/z of 1084 was detected. This measured mass differed by 1 Da from the mass measured in the corresponding peak of the O121 wild type trace eluting at 58.8 min. Likewise an m/z of 1042 was measured for the 2AB-labeled molecule with a retention time of 73.5 min. CID MS-MS of these precursor ions (Fig. 3c and d) resulted in a fragmentation pattern that resembled the spectra obtained from the O121 wild type 2AB-labeled glycans. The measured mass difference of 1 Da was assigned to residue c of the glycan structure. The mass difference of 1 Da corresponded to the calculated mass difference between an acid and an amide group, in agreement with the published structure of the wbqG mutant O antigen [23].
Furthermore polymerized 2AB-labeled O antigen subunits were identified in the O121 wild type trace. Two subunits variably O-acetylated were identified in the peak fractions with the retention times 83.1 min, 86.9 min and 90.6 min, respectively. The double acetylated species eluted first followed by the single acetylated and non-acetylated form. Due to the separation of the acetylated and non-acetylated forms, the degree of O-acetylation could be determined. In both strains approximately 50 % of the single repeating units were O-acetylated.
Precursors of the peptidoglycan monomer were also identified in some peaks of the O121 wild type trace (Fig. 2b). Peptidoglycan precursors are also assembled on Und-PP and are expected to be purified and labeled with the method used for O antigen subunits.
Production of glycoconjugates
The structure of the O121 wbqG mutant was confirmed, and it was shown to be cross-reactive with antibodies raised against the Vi antigen. Next, we examined whether the wbqG mutant O polysaccharide can elicit antibodies that bind to the Vi. Therefore, glycoconjugates were prepared for immunization studies. Glycoproteins were produced by expressing the bacterial oligosaccharyltransferase PglB, the engineered periplasmic carrier protein EPA, and either the E. coli O121 wild type or the wbqG mutant antigen in the E. coli K12 derivative CLM24 [16]. Strain CLM24 lacks the O antigen ligase (WaaL). Therefore, the transfer of O antigen to lipid A-core is blocked and the Und-PP linked O antigen substrate accumulates at the periplasmic face of the inner membrane providing the O antigen donor for the PglB-catalyzed transfer to specific asparagine residues within the protein acceptor. Additionally, E. coli K12 derivatives lack a functional endogenous O antigen gene cluster [24, 25]. A plasmid encoded O antigen gene cluster can therefore be expressed without producing mixed O antigen populations.
As described elsewhere EPA was used as protein acceptor with a N-terminal signal sequence for Sec-dependent secretion to the periplasm, and a C-terminal hexahistidine tag for purification by affinity chromatography [26]. Furthermore, EPA contained two engineered N-glycosylation sites. The low copy plasmid pGVXN114 was used for the expression of PglB under the control of the IPTG inducible tac promoter.
After induction of PglB and EPA, the newly synthesized glycoprotein was purified from periplasmic extracts by nickel affinity chromatography. Due to the presence of negatively charged polysaccharides in the glycoconjugate, anion exchange chromatography was used to separate the glycosylated from the non-glycosylated forms. Based on the separation of the two species it was found that in cultures expressing the O121 wild type O polysaccharide gene cluster approximately 70 % of the total EPA was glycosylated. The glycosylation efficiency was lower in cultures expressing the wbqG mutant O antigen whereas 35 % of the total carrier protein contained the glycan modification.
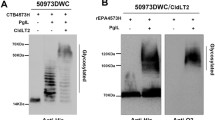
The purified glycoconjugates were separated by SDS-PAGE and visualized by Coomassie blue staining or by western blot after transfer to a nitrocellulose membrane using anti EPA, anti O121, and anti Vi antibodies (Fig. 4). By Coomassie blue staining a band of the same mass as that of unglycosylated EPA (70 kDa) could be detected in the purified O121 polysaccharide-EPA conjugate (O121-EPA), that is also recognized by the anti EPA but not the anti O121 sera. Therefore, unglycosylated EPA was largely removed in the glycoconjugate preparations. Mainly, a ladder of bands clustered between 100 and 130 kDa was detected by Coomassie blue staining. These bands reacted with anti-EPA serum, indicating modified forms of EPA. These larger polypeptides, but not EPA modified with the Shigella dysenteriae O1 antigen (O1-EPA) (described in [26]), were also detected with anti-O121 specific antibodies indicating the modification of the carrier with the co-expressed polysaccharide. EPA glycosylated with the wbqG mutant O polysaccharide (O121 wbqG -EPA) was additionally stained with anti Vi antibodies.
Production of glycoconjugates using the bacterial N-glycosylation system. Glycoconjugates were produced in E. coli CLM24 by co-expressing the bacterial oligosaccharyltransferase PglB, the engineered carrier protein EPA, and genes driving the synthesis of an antigenic polysaccharide (E. coli O121, E. coli O121 wbqG mutant, Shigella dysenteriae O1). Purified glycoconjugates were analyzed by SDS-PAGE, followed by Coomassie blue staining or by western blot after transfer to nitrocellulose membranes using anti EPA, anti O121, and anti Vi antibodies
As determined by SDS-PAGE analysis, mainly mono-glycosylated EPA was purified, i.e. EPA modified on one of the two engineered glycosylation sites with the corresponding O polysaccharide. Traces of di-glycosylated EPA could be detected by western blot in the purified O121 wbqG -EPA sample (Fig. 4). The di-glycosylated form of EPA runs as a second fainter ladder of bands slightly bigger than 130 kDa. As seen in Fig. 2a, the expressed O antigens display a modal chain length distribution with an average of 12 repeating units. Assuming the purified glycoconjugates consisted of mono-glycosylated EPA, containing a single polysaccharide chain of an average length of 12 repeating units, the sugar-to-protein weight ratio was estimated to be 0.15:1.
Immunogenicity of the glycoconjugates in mice and evaluation of the polysaccharide specific antibody response
Next we analyzed the immune response elicited in mice upon immunization with the conjugate vaccines. Pilot experiments were conducted in small groups of CB6F1 mice to determine the dose range and adjuvantation of the purified glycoconjugates. These established that 20 μg of protein (approximately 3 μg of polysaccharide), in combination with Alum, were reproducibly immunogenic (data not shown). Subsequently, groups of CB6F1 mice (7 per group) were immunized subcutaneously on days 1, 22 and 57 with O121-EPA, O121 wbqG -EPA, or with 5 μg of purified Vi polysaccharide (Typhim Vi, Sanofi Pasteur MSD). Mice were sample bled on days 32 and 67 and the sera were tested for the presence of anti O 121 LPS and anti Vi total immunoglobulin (Ig). By day 67, a significant rise in serum Ig anti O121 LPS titer was observed in 13 of 14 animals immunized with either conjugate (Fig. 5a). One animal in the group of mice that were immunized with O121 wbqG -EPA did not show seroconvertion. Interestingly, the same animal developed a significant rise in serum Ig anti-Vi titer (Fig. 5b). As expected, the control group that was immunized with purified Vi polysaccharide did not show a detectable anti-O121 LPS response but a significant rise in serum Ig anti-Vi titer.
Immunization studies with glycoconjugates. Groups of mice were immunized with purified glycoconjugates adjuvanted with Alum. The control group was immunized with purified Vi polysaccharide. a Anti O121 total immunoglobulin titers of sera collected on day 67. b Anti Vi antibody titer titers of sera collected on day 67. Data is represented as individual (black circle) and mean (–) titers. One animal immunized with the O121 wbqG -EPA conjugate did not develop an O121-LPS specific antibody response, but the same animal showed a significant rise in anti-Vi antibody titer
Discussion
In this study a novel method for the analysis of Und-PP linked glycans is presented. The procedure described here is based on the method used to analyze dolichyl pyrophosphate (Dol-PP)-linked oligosaccharides of eukaryotic cells. Main modifications include an optimized extraction procedure for bacterial glycolipids and a purification step prior to glycan release by mild acid hydrolysis. The purification strategy of bacterial Und-PP-linked glycans is further complicated by the vast variety of different sugar structures assembled on this lipid carrier. The choice of an appropriate expression strain used to analyze a specific subclass of Und-PP-linked glycans is crucial. In this report, Und-PP-linked O polysaccharides were analyzed. Since Und-PP-linked O antigens represent an intermediate species of LPS biosynthesis, an E. coli strain was used lacking the O antigen ligase (ΔwaaL). Therefore, Und-PP-linked O polysaccharides are not transferred to lipid A-core, resulting in accumulations of this lipid intermediate. If O antigens were expressed in a waaL positive strain no 2AB-labeled O glycans could be identified, most likely due to the rapid turnover of this glycolipid species. Furthermore, O antigens are polymerized structures with high molecular weights, making it increasingly difficult for analysis by mass spectrometry. We therefore chose a strain background containing a mutation in the O antigen chain length regulator (wzz) gene involved in efficient polymerization of O antigen subunits. This resulted in the production of mainly single repeat units and short polymerized O antigens, hence simplifying MS analysis. As mentioned previously, several other polysaccharide structures are also assembled on Und-PP, like peptidoglycan precursors, capsular polysaccharides and the enterobacterial common antigen (ECA), which might complicate the identification and characterization of O glycan species. We therefore used E. coli strain SCM6 for O antigen expression, which contains deletions in all major polysaccharide gene clusters.
With this modified method the O121 wbqG mutant O polysaccharides was analyzed. This study confirms the published structure by King et al. [23]. Furthermore, we could show that the recombinantly expressed wbqG mutant O antigen structure contained O-acetylated N-acetylgalactosaminuronic acid, most likely modified at C-3. Therefore this mutant O polysaccharide contains structural motifs also present in the Vi. O-acetyl groups of the Vi polysaccharide form an immunodominant epitope and immunogenicity of Vi is closely related to the degree of O-acetylation [19, 20].
For the first time it is reported that the wbqG mutant O polysaccharide is cross-reactive with antibodies raised against the Vi antigen. The strategy of using cross-reactive polysaccharide structures as vaccine components has been evaluated in several published studies. For example the cell wall polysaccharide (CWP) of the nonpathogenic bacterium Bacillus pumilus strain Sh18 was shown to be cross-reactive with the capsular polysaccharide (CPS) of Haemophilus influenzae type b. Conjugates containing the Sh18 CWP induced antibodies that reacted in an enzyme-linked immunosorbent assay (ELISA) with the CPS of H. influenzae type b strains [27]. Another study reported that Shewanella spp. CPSs share structural features with glycoproteins found in the Bacillus anthracis spore. A conjugate containing the Shewanella CPS induced antibodies that bound to B. anthracis spores and it will be further evaluated as a component of an anthrax vaccine [28].
Similarly, glycoconjugates composed of the E. coli O121 wild type or the wbqG mutant O polysaccharide and the P. aeruginosa exotoxin A (O121-EPA/O121 wbqG -EPA) were prepared in this study. EPA has already been successfully used as immunogenic carrier in a typhoid conjugate vaccine [12]. Both groups of mice immunized with glycoconjugates developed glycan specific antibody responses. 6 of 7 mice immunized with the O121 wbqG -EPA conjugate showed a significant rise in serum immunoglobulin (Ig) anti-O121 LPS titer, indicating that other antigenic determinants than the uronamide groups are important for inducing an anti-O121 LPS specific immune response. Interestingly, antibodies of one animal immunized with the O121 wbqG -EPA conjugate were not reactive with the E. coli O121 LPS but rather with the Vi polysaccharide. This implies that this animal developed an antibody response against the epitope constituted by residues b and c’ (Fig. 1), which resembles the Vi structure. However, the other animals of this group raised antibodies against an O121-LPS specific epitope, most likely residue d, containing a prominent surface exposed side group. Further optimizations of the O121 glycan structure might improve a Vi specific immune response upon immunization and experiments are planned to investigate if immunization with the O121 wbqG -EPA conjugate primes a Vi-specific immune response that could be boosted with purified Vi polysaccharide.
Experimental procedures
Bacterial strains, plasmids, and culture conditions
All bacterial strains and plasmids used in this study are listed in Table 1. Construction of the plasmids is described below. E. coli strains were grown in LB medium (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) or LB agar (LB medium with the addition of 15 g agar per liter) at 37 °C. S. Typhi BRD948 was grown in LB medium supplemented with 1 % v/v Aro-mix (40 mg L-phenylalanine, 40 mg L-tryptophan, 10 mg 4-aminobenzoic acid, and 10 mg 2,3-dihydroxybenzoic acid in 10 ml of ddH2O) and 1 % v/v Tyr-mix (40 mg L-tyrosine disodium salt in 10 ml ddH2O) at 37 °C. If appropriate, the media contained tetracycline (20 μg ml−1), spectinomycin (80 μg ml−1), or ampicilin (100 μg ml−1).
DNA manipulations
Plasmid DNA was isolated using the NucleoSpin Plasmid or NucleoBond Xtra Maxi Plus kit (Macherey-Nagel). Total chromosomal DNA was isolated using NucleoSpin Tissue kit (Macherey-Nagel). Restriction enzymes (Fermentas), shrimp alkaline phosphatase (Fermentas), T4 DNA ligase (Fermentas), and Phusion High-Fidelity DNA polymerase (Finnzyme) were used according to the manufacturer’s instructions. PCR and restriction fragments were purified for cloning using the NucleoSpin Extract II kit (Macherey-Nagel). All DNA sequencing was completed by Synergene Biotech GmbH (Switzerland) and synthetic oligonucleotides were ordered at Microsynth AG (Switzerland).
Plasmid constructions
pGVXN157 contains a synthetic oligonucleotide cassette formed from annealing of 5′- AATTGGCGCGCCCGGGACTAGTCTTGGG and 5′- AATTCCCAAGACTAGTCCCGGGCGCGCC ligated into the EcoRI-digested pLAFR1 [29], thereby introducing unique AscI and SpeI single restriction sites. The E. coli O121 O antigen cluster was amplified from genomic DNA prepared from E. coli O121 (CCUG 11422) using the primers 5′- AAAGGCGCGCCGCGAAGGTAAAGTCAGCCG and 5′- AAAACTAGTCAGGAGTGAATTAAGTCATTG. The digested PCR fragment was ligated into the AscI/SpeI digested pGVXN157 resulting in pGVXN331. pGVXN333 was constructed by inserting a synthetic oligonucleotide cassette formed from annealing of 5′- TGAATGAATGAACTAGTTCAATCACTCA and 5′- TGAGTGATTGAACTAGTTCATTCATTCA into the single restriction site PmlI, interrupting the open reading frame of wbqG.
LPS analysis
Cells of an overnight culture equivalent to an A600 of 1 were collected, resuspended in 100 μl of 1 × sample buffer according to Laemmli [30] and boiled at 95 °C for 10 min. Proteinase K (Fermentas) was added to a final concentration of 200 μg/ml and the sample was incubated at 60 °C for 1 h. The LPS molecular species from the proteinase K-digested whole cell lysates were separated by SDS-PAGE using a 12 % BisTris NuPAGE gel from Invitrogen and MES running buffer according to manufacturer’s instructions. LPS was visualized by staining with silver [31]. Immunological properties of O antigens were analyzed by Western blot using standard methods. The structure of the E. coli O121 O antigen is identical to the Shigella dysenteriae type 7 O antigen therefore an anti S. dysenteriae type 7 sera was purchased from Reagensia AB (Sweden) and used in a 1:100 dilution. Anti Vi polyclonal antibody was purchased from Murex Biotech Ltd (England) and used in a 1:100 dilution.
Analysis of Und-PP linked O antigen glycans
The O antigen glycans were analyzed in E. coli strain SCM6 (C. Marolda and M. Valvano, unpublished), which contains chromosomal deletions in several polysaccharide gene clusters. The O polysaccharide was expressed by transforming SCM6 cells with a plasmid encoding the O antigen cluster and the wecA expression plasmid pGVXN121. SCM6 transformed with empty plasmids was used as a negative control to identify O antigen specific signals. The strains were grown over night in a shake flask. Cells equivalent to an A600 of 400 were harvested, washed once with 0.9 % NaCl, and lyophilized. Lipids were extracted from the dried cells with 95 % methanol (MeOH) by repeated rounds of vortexing and incubation on ice for 10 min. The suspension was converted into 85 % MeOH by the addition of ddH2O and further incubated for 10 min on ice while regularly vortexing. After centrifugation, the supernatant was collected and the extract was dried under N2. The dried lipids were dissolved in 1:1 methanol/water (M/W) containing 10 mM tetrabutylammonium phosphate (TBAP) and subjected to a C18 SepPak cartridge (Waters Corp., Milford, MA). The cartridge was conditioned with 10 ml MeOH, followed by equilibration with 10 ml 10 mM TBAP in 1:1 M/W. After loading of the sample, the cartridge was washed with 10 ml 10 mM TBAP in 1:1 M/W and eluted with 5 ml MeOH followed by 5 ml 10:10:3 chloroform/methanol/water (C/M/W). The combined elution fractions were dried under N2.
The lipid sample were hydrolyzed according to Glover et al. [32] by dissolving the dried samples in 2 ml 1 M trifluoroacetic acid (TFA) in 50 % n-propanol and heating to 50 °C for 15 min. The hydrolyzed sample was dried under N2, dissolved in 4 ml 3:48:47 C/M/W and subjected to a C18 SepPak cartridge (Waters Corp., Milford, MA) to separate the lipids from the hydrolyzed glycans. The cartridge was conditioned with 10 ml MeOH, followed by equilibration with 10 ml 3:48:47 C/M/W. The sample was applied to the cartridge and the flow-through was collected. The cartridge was washed with 4 ml 3:48:47 C/M/W and the combined flow-through fractions were dried using a SpeedVac.
The dried samples were labeled with 2-aminobenzamide (2AB) according to Bigge et al. [33]. The glycan clean-up was performed using the paper disk method as described in Merry et al. [34]. The separation of 2AB-labeled glycans was performed by HPLC using a GlycoSep N normal phase column according to Royle et al. [35], but modified to a three solvent system. Solvent A: 10 mM ammonium formate pH 4.4 in 80 % acetonitrile. Solvent B: 30 mM ammonium formate pH 4.4 in 40 % acetonitrile. Solvent C: 0.5 % formic acid. The column temperature was 30 °C and 2AB-labeled glycans were detected by fluorescence (λex = 330 nm, λem = 420 nm). Gradient conditions: A linear gradient of 100 % A to 100 % B over 160 min at a flow rate of 0.4 ml min−1, followed by 2 min 100 % B to 100 % C, returning to 100 % A over 2 min and running for 15 min at 100 % A at a flow rate of 1 ml min−1, then returning the flow rate to 0.4 ml min−1 for 5 min. Samples were injected in ddH2O.
To identify O antigen specific glycans, the 2AB glycan profile from cells carrying an empty plasmid control was subtracted from the trace obtained from cells harbouring an O antigen cluster. The O antigen specific peaks were collected and 2AB glycans were analyzed on a MALDI SYNAPT HDMS Q-TOF system (Waters Corp., Milford, MA). Samples were dissolved in 5:95 acetonitrile/water and spotted 1:1 with 20 mg ml−1 DHB in 80:20 methanol/water. Calibration was done with PEG (Ready mixed solution, Waters Corp., Milford, MA), spotted 1:3 with 5 mg ml−1 α-cyano-4-hydroxycinnamic acid (CHCA, Sigma-Aldrich, Switzerland) in 60:40:0.1 acetonitrile/water/trifluoroacetic acid. The instrument was equipped with 200 Hz solid state UV laser. Mass spectra were recorded in positive ion mode. For MS/MS, laser energy was fixed at 240 at a firing rate of 200 Hz, collision gas was argon. A collision energy profile was used to ramp collision energy depending on the m/z. Combined, background subtracted, and smoothened (Savitzsky Golay) spectra were centered using MassLynx v4.0 software (Waters Corp., Milford, MA).
Production and purification of glycoconjugates
The production of glycoconjugates was achieved by expressing the oligosaccharyltransferase PglB, the engineered acceptor protein EPA, and a gene cluster producing undecaprenyl-pyrophosphate (Und-PP)-linked glycans in E. coli. pGVXN114 (expressing PglB), pGVXN150 (expressing C-terminal His6-tagged EPA) and pGVXN331 (O121 antigen cluster) or pGVXN333 (O121 wbqG mutant antigen) were co-transformed into E. coli strain Clm24 [16]. Cells were cultured in LB medium supplemented with antibiotics at 37 °C in the shaker incubator (180 rpm). Shake flask expression cultures were inoculated from an uninduced overnight culture to an A600 of 0.05. Expression of PglB and the carrier protein EPA was induced at an A600 of 0.4–0.6 by IPTG (1 mM) and L-arabinose (0.02 % w/v). Four hours after the first induction a second pulse of L-arabinose (0.02 % w/v) was added. Cells were harvested after overnight incubation (total induction time of 19–22 h). Pellets were washed with 0.9 % NaCl and suspended in resuspension buffer (25 % sucrose, 10 mM EDTA, 200 mM Tris HCl pH 8.0) at a concentration equivalent to an A600 of 50. The cell suspension was incubated on a shaker for 20 min at 4 °C. After centrifugation the cell pellet was resuspended in the same volume of osmotic shock buffer (10 mM Tris HCl pH 8.0). The suspension was incubated on a shaker for 30 min at 4 °C and centrifuged at 10,000 g for 20 min to remove the spheroblasts. The supernatant containing periplasmic proteins was collected and the recombinant EPA containing a C-terminal hexahistidine tag was purified using a HisTrap crude FF 1 ml column (GE Healthcare, Switzerland). The extract was diluted with 5 × HT binding buffer (2.5 M NaCl, 150 mM Tris HCl pH 8.0, 50 mM imidazole) to optimize the binding conditions and MgCl2 was added to a final concentration of 50 mM. The extract was filtered and applied to the HisTrap crude FF column equilibrated with 1 × HT binding buffer. After loading the column was washed with the same buffer containing 20 mM imidazole to remove unbound proteins. Proteins were eluted from the column with HT elution buffer (HT binding buffer containing 0.5 M imidazole).
Subsequently, the glycoprotein was separated from the unglycosylated EPA using a Resource Q 1 ml column (GE Healthcare, Switzerland). The HisTrap elution fractions containing EPA were pooled and diluted 10 × with RQ binding buffer (20 mM L-histidine, pH 6.0). The diluted EPA sample was applied to the anion exchange column equilibrated with RQ binding buffer. The column was eluted with a linear gradient from 0 % to 32.5 % of RQ elution buffer (RQ binding buffer containing 1 M NaCl) in 25 column volumes and 0.5 ml fractions were collected using an Äkta FPLC (Amersham Biosciences). The fractions were analyzed by SDS-PAGE and proteins were stained with Coomassie blue. Fractions containing glycoprotein were pooled and buffer was exchanged to PBS using an Amicon Ultra-4 centrifugal filter unit with a 30 kDa membrane (Millipore) by performing several concentration and dilution steps according to manufacturer’s instructions. The concentration of the final purified protein sample was adjusted to 1 mg ml−1.
Purification of E. coli O121 LPS
LPS of an E. coli O121 (CCGU 11422) culture was purified by phenol extraction as described elsewhere [36].
Purification of Vi polysaccharide and modification with tyramine
Vi polysaccharide was purified from S. Typhi BRD948 by a modified procedure as previously described [37]. Briefly, S. Typhi BRD948 was grown in LB medium supplemented with Aro- and Tyr-mix. After overnight incubation at 37 °C in the shaker incubator (180 rpm) the culture was heated to 60 °C for 1 h and centrifuged. Vi was precipitated from the supernatant with 0.1 % hexadecyltrimethylammonium bromide (CTAB, Sigma, H6269). 20 g l−1 celite 545 (Sigma, 20199-U) was added and the mixture was stirred for 1 h at room temperature (RT) in order to allow the formation of a polysaccharide-CTAB complex, which adsorbs onto the celite. The celite was poured into a reservoir of appropriate size (Extract-clean EV SPE Reservoir, Socochim S.A.) equipped with a frit (Socochim S.A.). The column was washed successively by gravity flow with 10 column volumes (CV) of 0.05 % CTAB, 10 CV of 20 % ethanol, 50 mM sodium phosphate buffer pH 6.0, and 14 CV of 45 % ethanol to eliminate adsorbed impurities. The Vi polysaccharide was finally eluted with 1.5 CV of 50 % ethanol, 0.4 M NaCl. Following elution, the polysaccharide was precipitated by the addition of ethanol to a final concentration of 80 % and incubation for 20 min at RT. Finally, the precipitated polysaccharide was collected by centrifugation for 20 min at 15,000 g, washed twice with 80 % ethanol, and lyophilized.
The protein and nucleic acid content of the purified Vi polysaccharide was determined by the bicinchoninic acid assay (BCA) and UV spectroscopy respectively. O-acetyl content was measured with acetylcholine as standard [38].
To increase the binding efficiency of the Vi to microtiter plates, the polysaccharide was tyraminated (Vi-Tyr). Tyramine hydrochloride (30 mg ml−1, Sigma) was added to 10 mg of purified Vi. 100 μl of 0.5 M N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide HCl (Sigma) was added and the mixture was incubated at pH 4.9–5.1 for 3 h. The reaction mixture was dialyzed against ddH2O.
Immunization studies
Groups of 7 CB6F1 female mice, 6–8 weeks old, were used in immunization experiments. Mice were immunized, subcutaneously, with 20 μg of glycoconjugate with Alum (Rehydragel LV- Aluminium Hydroxide, General Chemical) as adjuvant or 5 μg of Vi polysaccharide (Typhim Vi, Sanofi Pasteur MSD). Adjuvantation of the glycoconjugate was done just before immunization. Briefly, the purified glycoconjugates were diluted with PBS to a final concentration of 200 μg ml−1, Alum (final amount of Al3+ corresponded to 0.6 mg ml−1) was added, and the solution was gently mixed for 1 h at room temperature. Immunizations were performed on days 1, 22 and 57. Groups of mice normally received 100 μl doses of vaccines, corresponding to 20 μg of conjugate (protein). Blood samples were collected 10 days after the second and 10 days after the last immunization.
Enzyme-linked immunosorbent assay (ELISA) for murine antibodies
Flat bottom 96 well micro-titer plates (Nunc immuno PolySorb) were coated with 50 μl of 5 μg ml−1 E. coli O121 LPS or 5 μg ml−1 of tyraminated Vi (Vi-Tyr), diluted in PBS, at 4 °C overnight. The coating solution was poured away and the plate was submerged and vigorously agitated in 4,000 ml of wash buffer (1 × PBS with 0.05 % Triton × 100). This washing step was performed at least 4 times. Subsequently, the plate was dried by placing and spinning upside down in a micro plate rotor. This washing procedure was always applied in further washing steps. Each well was completely filled with 300 μl of blocking buffer (1 × PBS with 2.5 % BSA (globulin free BSA, Sigma, A7030)) and incubated 2 h at room temperature (RT) on a plate shaker. After washing and drying the plate, dilutions of mouse serum in dilution buffer (1 × PBS with 0.5 % BSA) were added to the plate (100 μl) and incubated 1 h at RT on a plate shaker. To detect total immunoglobulin (Ig), 100 μl of horseradish peroxidase (HRP) labeled goat anti-mouse Ig (Sigma) diluted 1:2000 in dilution buffer was added to each well and the plate was incubated for 1 h at RT on a plate shaker. Following washing and drying the plate, the reaction was developed with 100 μl of Ultra TMB substrate (3,3′,5,5′-tetramethybenzidine liquid substrate, Pierce) for 15 min and stopped with the addition of 100 μl of 2 M sulfuric acid. Optical density (OD) was measured at 450 nm.
To determine the endpoint titer a 95 % confidence level was defined according to [39]. As negative sample a pool of preimmune sera was used.
References
Crump, J.A., Luby, S.P., Mintz, E.D.: The global burden of typhoid fever. Bull. World Health Organ 82(5), 346–353 (2004)
Bhan, M.K., Bahl, R., Bhatnagar, S.: Typhoid and paratyphoid fever. Lancet 366(9487), 749–762 (2005)
Mirza, S.H., Beeching, N.J., Hart, C.A.: Multi-drug resistant typhoid: a global problem. J. Med. Microbiol. 44(5), 317–319 (1996)
Hone, D.M., Attridge, S.R., Forrest, B., Morona, R., Daniels, D., LaBrooy, J.T., Bartholomeusz, R.C., Shearman, D.J., Hackett, J.: A galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect. Immun. 56(5), 1326–1333 (1988)
Levine, M.M., Ferreccio, C., Black, R.E., Germanier, R.: Large-scale field trial of Ty21a live oral typhoid vaccine in enteric-coated capsule formulation. Lancet 1(8541), 1049–1052 (1987)
Black, R.E., Levine, M.M., Ferreccio, C., Clements, M.L., Lanata, C., Rooney, J., Germanier, R.: Efficacy of one or two doses of Ty21a Salmonella typhi vaccine in enteric-coated capsules in a controlled field trial. Chilean Typhoid Committee. Vaccine 8(1), 81–84 (1990)
Murphy, J.R., Grez, L., Schlesinger, L., Ferreccio, C., Baqar, S., Munoz, C., Wasserman, S.S., Losonsky, G., Olson, J.G., Levine, M.M.: Immunogenicity of Salmonella typhi Ty21a vaccine for young children. Infect. Immun. 59(11), 4291–4293 (1991)
Levine, M.M., Ferreccio, C., Cryz, S., Ortiz, E.: Comparison of enteric-coated capsules and liquid formulation of Ty21a typhoid vaccine in randomised controlled field trial. Lancet 336(8720), 891–894 (1990)
Levine, M.M., Ferreccio, C., Abrego, P., Martin, O.S., Ortiz, E., Cryz, S.: Duration of efficacy of Ty21a, attenuated Salmonella typhi live oral vaccine. Vaccine 17(Suppl 2), S22–S27 (1999)
Weintraub, A.: Immunology of bacterial polysaccharide antigens. Carbohydr. Res. 338(23), 2539–2547 (2003)
Landy, M.: Studies on Vi antigen. VI. Immunization of human beings with purified Vi antigen. Am. J. Hyg. 60(1), 52–62 (1954)
Szu, S.C., Taylor, D.N., Trofa, A.C., Clements, J.D., Shiloach, J., Sadoff, J.C., Bryla, D.A., Robbins, J.B.: Laboratory and preliminary clinical characterization of Vi capsular polysaccharide-protein conjugate vaccines. Infect. Immun. 62(10), 4440–4444 (1994)
Lin, F.Y., Ho, V.A., Khiem, H.B., Trach, D.D., Bay, P.V., Thanh, T.C., Kossaczka, Z., Bryla, D.A., Shiloach, J., Robbins, J.B., Schneerson, R., Szu, S.C.: The efficacy of a Salmonella typhi Vi conjugate vaccine in two-to-five-year-old children. N. Engl. J. Med. 344(17), 1263–1269 (2001)
Young, N.M., Brisson, J.R., Kelly, J., Watson, D.C., Tessier, L., Lanthier, P.H., Jarrell, H.C., Cadotte, N., St Michael, F., Aberg, E., Szymanski, C.M.: Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium. Campylobacter jejuni. J. Biol. Chem. 277(45), 42530–42539 (2002)
Wacker, M., Linton, D., Hitchen, P.G., Nita-Lazar, M., Haslam, S.M., North, S.J., Panico, M., Morris, H.R., Dell, A., Wren, B.W., Aebi, M.: N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science (New York, N.Y) 298(5599), 1790–1793 (2002)
Feldman, M.F., Wacker, M., Hernandez, M., Hitchen, P.G., Marolda, C.L., Kowarik, M., Morris, H.R., Dell, A., Valvano, M.A., Aebi, M.: Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 102(8), 3016–3021 (2005)
Wacker, M., Feldman, M.F., Callewaert, N., Kowarik, M., Clarke, B.R., Pohl, N.L., Hernandez, M., Vines, E.D., Valvano, M.A., Whitfield, C., Aebi, M.: Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc. Natl. Acad. Sci. U. S. A. 103(18), 7088–7093 (2006)
Kowarik, M., Young, N.M., Numao, S., Schulz, B.L., Hug, I., Callewaert, N., Mills, D.C., Watson, D.C., Hernandez, M., Kelly, J.F., Wacker, M., Aebi, M.: Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25(9), 1957–1966 (2006)
Szu, S.C., Li, X.R., Stone, A.L., Robbins, J.B.: Relation between structure and immunologic properties of the Vi capsular polysaccharide. Infect. Immun. 59(12), 4555–4561 (1991)
Szu, S.C., Bystricky, S.: Physical, chemical, antigenic, and immunologic characterization of polygalacturonan, its derivatives, and Vi antigen from Salmonella typhi. Methods Enzymol. 363, 552–567 (2003)
Whitfield, C.: Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 (2006)
Parolis, H., Parolis, L.A., Olivieri, G.: Structural studies on the Shigella-like Escherichia coli O121 O-specific polysaccharide. Carbohydr. Res. 303(3), 319–325 (1997)
King, J.D., Vinogradov, E., Tran, V., Lam, J.S.: Biosynthesis of uronamide sugars in Pseudomonas aeruginosa O6 and Escherichia coli O121 O antigens. Environ. Microbiol. 12(6), 1531–1544 (2010)
Liu, D., Reeves, P.R.: Escherichia coli K12 regains its O antigen. Microbiology 140(Pt 1), 49–57 (1994)
Feldman, M.F., Marolda, C.L., Monteiro, M.A., Perry, M.B., Parodi, A.J., Valvano, M.A.: The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274(49), 35129–35138 (1999)
Ihssen, J., Kowarik, M., Dilettoso, S., Tanner, C., Wacker, M., Thony-Meyer, L.: Production of glycoprotein vaccines in Escherichia coli. Microb. Cell. Fact. 9, 61 (2010)
Kubler-Kielb, J., Coxon, B., Schneerson, R.: Chemical structure, conjugation, and cross-reactivity of Bacillus pumilus Sh18 cell wall polysaccharide. J. Bacteriol. 186(20), 6891–6901 (2004)
Kubler-Kielb, J., Vinogradov, E., Hu, H., Leppla, S.H., Robbins, J.B., Schneerson, R.: Saccharides cross-reactive with Bacillus anthracis spore glycoprotein as an anthrax vaccine component. Proc. Natl. Acad. Sci. U. S. A. 105(25), 8709–8712 (2008)
Friedman, A.M., Long, S.R., Brown, S.E., Buikema, W.J., Ausubel, F.M.: Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18(3), 289–296 (1982)
Laemmli, U.K., Favre, M.: Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80(4), 575–599 (1973)
Tsai, C.M., Frasch, C.E.: A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119(1), 115–119 (1982)
Glover, K.J., Weerapana, E., Imperiali, B.: In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U. S. A. 102(40), 14255–14259 (2005)
Bigge, J.C., Patel, T.P., Bruce, J.A., Goulding, P.N., Charles, S.M., Parekh, R.B.: Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal. Biochem. 230(2), 229–238 (1995)
Merry, A.H., Neville, D.C., Royle, L., Matthews, B., Harvey, D.J., Dwek, R.A., Rudd, P.M.: Recovery of intact 2-aminobenzamide-labeled O-glycans released from glycoproteins by hydrazinolysis. Anal. Biochem. 304(1), 91–99 (2002)
Royle, L., Mattu, T.S., Hart, E., Langridge, J.I., Merry, A.H., Murphy, N., Harvey, D.J., Dwek, R.A., Rudd, P.M.: An analytical and structural database provides a strategy for sequencing O-glycans from microgram quantities of glycoproteins. Anal. Biochem. 304(1), 70–90 (2002)
Apicella, M.A.: Isolation and characterization of lipopolysaccharides. Methods Mol. Biol. 431, 3–13 (2008)
Demil, P., D’Hondt, E., Hoecke, C.V.: Salmonella Typhi vaccine compositions. European Patent EP1107787 (2003)
Hestrin, S.: The reaction of acetylcholine and other carboxylic acid derivatives with hydroxylamine, and its analytical application. J. Biol. Chem. 180(1), 249–261 (1949)
Frey, A., Di Canzio, J., Zurakowski, D.: A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 221(1–2), 35–41 (1998)
Hone, D.M., Harris, A.M., Chatfield, S., Dougan, G., Levine, M.M.: Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine 9(11), 810–816 (1991)
Dykxhoorn, D.M., St Pierre, R., Linn, T.: A set of compatible tac promoter expression vectors. Gene 177(1–2), 133–136 (1996)
Acknowledgement
E. coli SCM6 was kindly provided by M. Valvano. We thank Susanne Dura und Sacha Keller for their technical assistance in the laboratory.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wetter, M., Kowarik, M., Steffen, M. et al. Engineering, conjugation, and immunogenicity assessment of Escherichia coli O121 O antigen for its potential use as a typhoid vaccine component. Glycoconj J 30, 511–522 (2013). https://doi.org/10.1007/s10719-012-9451-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-012-9451-9