Abstract
Using a focused glycan-gene microarray, we compared the glycosyltransferase (GT) and sulfotransferase gene expression profiles of human monocytes, dendritic cells (DCs) and macrophages (Mϕs), isolated or differentiated from the same donors. Microarray analysis indicated that monocytes express transcripts for a full set of enzymes involved in the biosynthesis of multi-multiantennary branched N-glycans, potentially elongated by poly-N-acetyl-lactosamine chains, and of mucin-type Core 1 and Core 2 sialylated O-glycans. Monocytes also express genes involved in the biosynthesis and modification of glycosaminoglycans, but display a limited expression of GTs implicated in glycolipid synthesis. Among genes expressed in monocytes (90 out of 175), one third is significantly modulated in DCs and Mϕ respectively, most of them being increased in both cell types relative to monocytes. These changes might potentially enforce the capacity of differentiated cells to synthesize branched N-glycans and mucin-type O-glycans and to remodel cell surface proteoglycans. Stimulation of DCs and Mϕs with lipopolysaccharide caused a general decrease in gene expression, mainly affecting genes found to be positively modulated during the differentiation steps. Interestingly, although a similar set of enzymes are modulated in the same direction in mature DCs and Mϕs, cell specific genes are also differentially regulated during maturation, a phenomenon that may sustain functional specificities. Validation of this analysis was provided by quantitative real-time PCR and flow cytometry of cell surface glycan antigens. Collectively, this study implies an important modification of the pattern of glycosylation in DCs and Mϕs undergoing differentiation and maturation with potential biological consequences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Glycosylation of proteins and lipids plays a crucial role in numerous biological processes including the regulation of immune and inflammatory responses (for reviews,[1–6]). During physiological conditions, glycans exert diverse functions on the immune system. By serving as ligands for glycan-binding proteins, such as classical adhesion molecules and lectins, they mediate immune cell differentiation, survival, adhesion, and trafficking [7–13]. During stress or infection, glycans also play a pivotal role by triggering or controlling immune cell signalling, migration, expansion and/or effector functions [10, 14–20]. Glycans exposed on the surface of professional antigen (Ag) presenting cells (APCs) are likely to be critical in many aspects of immune responses. They mediate host-pathogen interactions, influence their tropism and emigration and shape their biological functions after cell-to-cell contact. For instance, glycans play a part in the cross-talk between dendritic cells (DCs), the most potent APCs, and conventional T lymphocytes to modulate the strength and the quality of the acquired immune response [21]. Moreover, interactions of APCs with cells of the innate system, including natural killer cells, are supported by glycan/counter-receptor interactions (for review, [22]). More recently, a new concept has emerged showing that the production of glycolipids (glycosphingolipids, GSLs) by CD1d-expressing APCs is critical to activate Natural Killer T cells, a sub-population of innate/memory non-conventional T lymphocytes (for reviews, [23, 24]). So far, although differentiation and activation of APCs, including monocytes, DCs and macrophages (Mϕs), are accompanied by programmed remodelling of cell surface (glycosylated) molecules with potentially biologically important consequences, comparative analysis of the expression of genes involved in glycan biosynthesis (essentially glycosyltransferases, GTs) and modification (mainly sulfotransferases) has only been reported in DCs [25, 26].
The mononuclear phagocyte system is composed by monocytes, DCs and Mϕs, which contribute to tissue remodelling and homeostasis, inflammation and immune defence. Circulating monocytes, which constitute ~ 5–10% of peripheral blood leukocytes in humans, give rise to tissue-resident Mϕs as well as to other specialized cells such as osteoclasts and myeloid DCs [27–30]. Dendritic cells are critical in the induction, expansion and regulation of immune responses (for reviews, [31–34]). Immature DCs principally locate at sites of Ag entry, where they are poorly immunogenic but efficient at capturing Ags through receptor-mediated endocytosis, macropinocytosis and phagocytosis. Upon activation by inflammatory factors and/or microbial components, DCs undergo a complex process of maturation allowing their traffic to T-cell areas of lymphoid tissues through modification of adhesion molecule and chemokine receptor expression. Dendritic cell maturation is also associated with high surface expression of Ag presenting and co-stimulatory molecules and with secretion of chemokines and bioactive mediators such as immuno-stimulatory cytokines. These factors are important to the downstream activation of numerous bystander innate immune cells, such as Natural killer (T) cells, and to promote T and B cell stimulation, expansion and polarisation [31–34]. Although Mϕs are much less potent in inducing primary immune response relative to DCs, they are crucial players in innate/acquired responses, in particular in immediate early defence against bacterial infection. The most prominent functions of Mϕs are phagocytosis of intruding micro-organisms and necrotic/apoptotic cells, bactericidal activity and rapid secretion of pro-inflammatory cytokines (for review, [27]). Like DCs, activation/maturation of Mϕs is mediated by engagement of various innate sensors, including Toll-like receptors (TLRs) (for reviews, [35, 36]).
In the present study, we aimed to obtain general information relating to the expression of genes encoding GTs and sulfotransferases in human monocytes, DCs and Mϕs isolated or differentiated from the same donor. We also aimed to compare the expression profile of these genes in immature versus mature (immuno-stimulatory phenotype) DCs and Mϕs. To this end, we took advantage of a focused gene microarray (glycogene-chip v3, Consortium for Functional Glycomics) using the Affymetrix technology. The glyco-gene-chip v3 is a custom designed GeneChip expression array that utilizes 21 probes for each targeted transcript, ten more than are found on the commercially available GeneChip arrays such as the Hu133 Plus 2.0 array from Affymetrix. The additional probes used on this focused array increase the sensitivity for detection of low abundant transcripts (unpublished results, SRH). In the present study, we report that the differentiation and maturation processes affected the gene expression profiles of GTs and sulfotransferases in DCs and Mϕs. Potential functional consequences for these changes are discussed.
2 Materials and methods
2.1 Reagents
Human recombinant GM-CSF was purchased from Peprotech (Rocky Hill, NJ) and IL-4 were from R&D systems (Abingdon, UK). LPS (type 055B5) was purchased from Sigma-Aldrich (St Louis, MI). Anti-CD14-microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany) were used for magnetic cell separation. The FITC-conjugated mouse anti-CD1a, anti-CD16, anti-CD209 (DC-SIGN) and anti-HLA-DR mAbs were obtained from Becton Dickinson (San Diego, CA). The APC-conjugated anti-CD11c and anti-CD86 mAbs as well as the PE-conjugated mouse anti-CD80 and anti-CD14 mAbs were purchased by Becton Dickinson, whereas the anti-CD83 mAb was from Coulter (Miami, Flo). The anti-Mϕ mAb (clone RFD7) was furnished by AbD-Serotec (Abingdon, UK). The anti-GM3 (clone GMR6), anti-GD3 (clone S2-566), anti-Lea (clone 7LE), anti-Leb (clone 2-25 L), anti-Ley (clone H18A) and anti-Lewisx (Lex, clone 73-30) mAbs were from Seikagaku Corporation (Tokyo, Japan). The anti-sialyl Tn (clone HB-STn1), anti-sLea (clone 2D3) and anti-Tn Ag (clone M0896) were from Dako (Glostrup, Denmark) and the anti-sialyl Lex (sLex, clone CSLEX1) from Becton Dickinson. The anti-PSA Ab (clone 735) was kindly provided by Prof. R. Gerardy-Schahn (Medizinische Hochschule, Hannover, Germany). The FITC-conjugated Sambucus nigra agglutinin (SNA) lectin was from Vector Laboratories (Burlingame, CA).
2.2 Preparation and stimulation of human DCs and Mϕs
After giving informed consent, blood from healthy volunteers was collected (Etablissement Français du Sang, Lille). Peripheral blood mononuclear cells were prepared by density centrifugation on Ficoll-Paque. Blood monocytes were purified by positive selection over a MACS column using anti-CD14-conjugated microbeads. This purified cell population contained at least 95% CD14+ cells. An aliquot containing about 3–5 × 106 monocytes was immediately frozen to prepare RNA. Monocytes were then differentiated into DCs [37, 38] or into Mϕs [39] by standard procedures. Briefly, monocytes were cultivated at 106 cells/ml for 6 days in RPMI 1640 with 10% heat-inactivated FCS (Invitrogen, Paisley, UK) containing 10 ng /ml IL-4 and 25 ng /ml GM-CSF or GM-CSF alone to obtain myeloid DCs [40, 41] or proinflammatory type I Mϕs [42, 43], respectively. At day 3, half of the culture medium was renewed by addition of fresh complete medium containing cytokines. At day 5, DCs and Mϕs (at least 95% pure, as revealed by flow cytometry) were stimulated or not with LPS (100 ng/ml). Cells were collected after 18 h stimulation to prepare RNA or after 24 h for FACS analysis. Cell death was assessed by trypan blue exclusion and measurement of MTT oxydo-reduction (Sigma) in all culture conditions and neither exceeded 10%.
2.3 Microarray analysis of gene expression
Analysis of gene expression was conducted using a custom gene microarray (GLYCOv3 chip) produced by Affymetrix for the Consortium for Functional Glycomics (www.functionalglycomics.org), and containing probe sets for over 1,000 human genes including 199 human GTs and sulfotransferases. In this study, we focused our analysis on the expression of the 175 genes involved in the biosynthesis of N-glycans, mucin-type O-glycans, glycosaminoglycans, and glycolipids. Five to six independent experiments were performed for each condition. Total RNA was extracted using the Qiagen Mini kit according to the manufacturer’s suggested protocol (Qiagen, Inc., Valencia, CA). The quality of the samples was checked with an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). For GT and sulfotransferase expression analysis, total RNA (1 µg) was amplified and biotin-labelled using the Bioarray MessageAmp II-Biotin Enhanced Kit from Ambion and then hybridized to the GLYCOv3 array. Hybridization and scanning of samples was performed using standard Affymetrix protocols for GeneChip expression arrays based on methods originally described in [44] (protocol available at http://affymetrix.com). Chips were scanned using the Affymetrix ScanArray 3,000 using default settings and a target intensity of 250 for scaling. The transcriptional profile was evaluated in independent cell preparations, each derived from a different donor and one chip per biological sample was run. Expression calls indicate whether a gene is detected above background using the GC-matched bins as background measurement. For this purpose the Fisher’s Combined P Method [45] as implemented in the R program software (R is available as Free Software under the terms of the Free Software Foundation’s GNU General Public License). The Fisher’s Combined P method threshold was adjusted so that the expression calls were consistent with the present and absent calls as implemented in the Affymetrix Microarray Suite V5. Intensity values were normalized using Robust Multichip Average (RMA) [46, 47]. The Limma package in the R software was used for the Anova analysis. The fold changes and standard errors were estimated by fitting a linear model for each gene and empirical Bayes smoothing was applied to the standard errors for all the samples at the same time. Statistics were obtained for transcripts with the multiple testing adjusted (Benjamini-Hochberg) p-values to a level of 0.05. Filtering was performed so that probe-sets were eliminated from the results with a fold change of <1.4. Heatmaps were generated by dChip (http://biosun1.harvard.edu/complab/dchip/). Raw data files for each of the experiments performed are available at the consortium for Functional Glycomics website (www.functionalglycomics.org/fg) under Resource Request no. 268.
2.4 Gene expression analysis by qPCR
Changes in gene expression observed by microarray analyses were verified by qPCR for some genes. Briefly, DNA was synthesized from 1 μg of total RNA with random hexamer primers and Superscript reverse transcriptase (Invitrogen, Cergy Pontoise, France) using standard procedures. cDNAs were used as templates for PCR amplification using the SYBR® Green PCR Master Mix (Molecular Probes, Leiden, The Netherlands) and the ABI PRISM 7700 Sequence Detector (Applied Biosystems, Foster City, CA). Primers listed in Table 1 were designed by the Primer Express Program (Applied Biosystems) and used for amplification in triplicate assays. PCR amplification of GAPDH was performed to control for sample loading and to allow normalization between samples. ΔCt values were obtained by deducting the raw cycle threshold (Ct values) obtained for β-actin mRNA, the internal standard, from the Ct values obtained for investigated genes. Data are expressed as fold mRNA level increase or decrease compared to the mRNA expression level in non-differentiated (immature DCs or immature Mϕs vs. monocytes) cells or to non-stimulated cells (mature DCs or mature Mϕs vs. immature DCs or immature Mϕs, respectively).
2.5 Flow cytometry analysis
Cells were collected in PBS containing 2 mM EDTA at 4°C and labelled as previously described [37]. Cells were incubated for 30 min in aliquots of 2 × 105 cells in 50 µl of PBS containing 2% FCS with or without Abs (10 µg/ml) or the lectin SNA (40 µg/ml). After washing, cells were directly analyzed for the conjugated mAbs. In some cases, cells were incubated with Phycoerythrin-conjugated goat IgG anti-mouse IgG or IgM (Southern Biotechnology, Birmingham, Al). After 30 min incubation, labelled cells were washed and analyzed. Flow cytometry data were acquired on viability-gated cells using a FACSCalibur flow cytometer and analyzed with the CellQuest software system (BD Biosciences). Results are expressed as the mean fluorescence intensity (MFI) obtained with specific mAbs or lectins minus the value obtained with the isotype control (ΔMFI).
3 Results
3.1 Expression profile of GT and sulfotransferase genes in monocytes
We first analyzed the expression pattern of GT and sulfotransferase genes involved in the biosynthesis of N-glycans, mucin-type O-glycans, glycosaminoglycans (GAGs), and glycolipids in circulating peripheral blood monocytes. We found that, among the 175 analyzed genes (200 probe set), 90 are significantly expressed in human monocytes (five donors). Table 2 shows the average expression signals of these genes, which are clustered according to the specificities of the enzymes that they encode. As shown in Table 2, transcripts for fucosyltransferases (FucT), and to a lesser extent for glucosyltransferases (GlcT), glucuronosyltransferases (GlcAT) and mannosyltransferases (ManT) are expressed at a lower level compared to those of the other subfamilies. Interestingly, several genes involved in the first steps of N-glycosylation, in particular in the formation of the oligosaccharidyltransferase complex such as DAD1 (probes #92/93), RNP1 (#94), RNP2 (#95) and DDOST (#96), are highly expressed in monocytes. Concomitantly, genes encoding enzymes involved in both N-glycan branching (i.e. MGAT5B (#34), MGAT2 (#36), MGAT4B (#37/38/41) & MGAT1 (#40)) and poly-N-acetyl-lactosamine chain elongation (i.e. B3GNT1 (#29), B3GNT2 (#30), B3GNT8 (#35), IGNT2 (#31), B4GALT3 (#22) and B4GALT4 (#17)) are also well expressed, suggesting that monocytes synthesize N-glycans carrying di, tri, and tetra-antennary structures, possibly elongated by poly-N-acetyl-lactosamine chains. In parallel, monocytes also highly express B4GALT5 (#24) and B4GALT1 (#25), two genes encoding type 2 disaccharide (Galβ1-4GlcNAc) synthesising enzymes, together with fucosyltransferases (FUT4 (#6) and FUT7 (#5)) and sialyltransferase (ST3GAL6 (#58)) using type 2 disaccharide as acceptor substrate. This indicates that monocytes possess a full set of enzymes for the biosynthesis of Lex and sialyl (s)Lex in terminal position of oligosaccharide chains. In agreement with previous reports [48, 49], FACS analysis confirmed expression of sLex, but not Lex, on monocytes (Table 3). At the opposite, the β1,3-galactosyltransferases (β3-Gal T1, β3-Gal T2, β3-Gal T5 and β3-Gal T7) involved in the biosynthesis of type I disaccharide unit (Galβ1-3GlcNAc) are not significantly expressed in monocytes. This correlates with the absence of type I derived blood group Ags, such as Lea, sLea or Leb, as revealed by FACS analysis (Table 3). Polypeptide GalNAc-transferases, the mucin-type O-glycan initiating enzymes, are also well produced in monocytes. Seven different enzymes (i.e. ppGalNAc T3 (#7), T10 (#8/12), T7 (#9), T11 (#10), T2 (#11), T1 (#14), T6 (#15/16)) are fairly or highly expressed, suggesting a very good capacity for monocytes to synthesise O-glycan chains. In parallel, monocytes also express the Core 1 β3-Gal T1 (CIGALT1, #18), the Core 2 β6-GlcNAc T1 (GCNT1, #28) and several core 1-specific sialyltransferases (i.e. ST6GalNAc IV (#59), ST6GalNAc II (#61), and ST3Gal I (#65), but other core-specific enzymes such as Core 2 β6-GlcNAc T3, Core 2 and 4 β6-GlcNAc T4 or Core 3 β3-GlcNAc T6 are not detected. Moreover, monocytes do not express a transcript for ST6GalNAc I, the enzyme responsible for the synthesis of sialyl-Tn (NeuAcα2-6GalNAcα1-O-Ser/Thr). These data, together with the absence of Tn (GalNAcα1-O-Ser/Thr) or sialyl-Tn Ags (FACS analysis, data not shown), suggest that monocyte O-glycan chains are mainly Core 1 and Core 2 sialylated glycans. This is in line with recent data reporting mass spectrometry O-glycan profiling of monocytes [26]. Besides N-glycan and mucin-type O-linked glycan genes, monocytes express genes encoding for enzymes involved in GAG biosynthesis, including those implicated in the tetrasaccharide linker, chondroitin sulphate and heparan sulfate copolymerases, and for modification enzymes such as 5-epimerase and numerous sulfotransferases (Table 2). Finally, although the expression levels of the two key GTs controlling the first step of GSL synthesis (glucosylceramide synthase UGCG, #43) and lactosylceramide synthase) are low or undetectable, monocytes transcribe several genes encoding enzymes involved in GSL glycosylation. For instance, in the Globo series of GSLs, the Gb4 synthase (GBGT1, #13), but not the Gb3 synthase, is highly expressed in monocytes. In the Ganglio series, the GM3 synthase (ST3Gal V, #64) is highly expressed but both GD3 synthase (ST8Sia I), GT3 synthase (ST8Sia V) and GM2 synthase (β4-GalNAc T1) are not detected. Together, this indicates that ganglioside biosynthesis is rather limited to the a-series of gangliosides in monocytes.
3.2 Comparison of GT and sulfotransferase gene expression in DCs and Mϕs relative to monocytes
In humans, monocytes have the capacity to differentiate into CD11c+ MHC class II+ DCs in the presence of granulocyte Mϕ-colony stimulating factor (GM-CSF) and interleukin-4 or into Mϕs in the presence to GM-CSF alone [38, 50, 51]. We investigated whether in vitro differentiation of DCs or Mϕs from monocytes leads to changes in the level of GT and sulfotransferase transcript expression. The quality of the differentiated cells was assessed after selection and differentiation by flow cytometry. As shown in Table 3, clear phenotypic differences between monocytes, DCs and Mϕs was observed. Unlike Mϕs and monocytes, immature DCs express the canonical markers CD1a and DC-SIGN (CD209). At the opposite, Mϕs, but not DCs, produce CD16 and RFD7. Monocytes are strongly positive for CD14, a marker lost during their differentiation, and some cells are also positive for CD16. Furthermore, transcriptomic analysis confirmed that cell-specific markers are expressed by DCs, such as the lectins DEC-205, DC-SIGN (CD209) and DC immunoreceptor (DCIR), and by Mϕs, such as the scavenger receptors collectin 12 and LOX-1 or are common to both cell types such as the macrophage mannose receptor (CD206) (data not shown).
As shown in heatmap representation (Fig. 1), in both DCs and Mϕs, a significant number of GT and sulfotransferase genes (31/90 (34 %) and 27/90 (30 %), respectively) are significantly (P < 0.05) changed in their expression levels (fold change > 1.4). Indeed, 21 GT and sulfotransferase transcripts are increased and only ten are decreased in DCs, whilst in Mϕs, 22 GT and sulfotransferase mRNAs are increased and five are decreased. To validate these changes, we performed quantitative real-time PCR (qPCR) using biological samples (three to five) independent of those used in the gene chips analysis. To this end, we focused on genes with the highest expression variation and/or involved in specific pathways. As Table 4 shows, and among the tested genes, the qPCR analysis confirmed data obtained by the micro-array approach. Strikingly, DCs and Mϕs exhibit similarities in their pattern of GT and sulfotransferase transcript expression, indicating that the majority of these genes are modulated in the same direction during the differentiation processes (Fig. 1). Among them, several genes coding for enzymes involved in the first steps of N-glycan biosynthesis are increased (albeit moderately) in differentiated cells, such as those directly associated to the biosynthesis of the lipid-linked precursor of Asn-linked glycans, the dolichol-PP-oligosaccharide (ALG1 (#53), ALG2 (#54), ALG5 (#47), ALG8 (#46) and ALG9 (#51)) and to the transfer of the oligosaccharide onto the nascent protein (DAD1 (#93) and DDOST (#96)). Variation of gene transcripts are also observed for GTs that selectively act in the Golgi processing of N-glycans, such as the N-acetyl-glucosaminyltransferase MGAT2 (#36), MGAT4A (#27), and particularly the β3-GlcNAc T1 (# 29), the latter being essential for the synthesis of poly-N-acetyllactosamine chains [52]. These enzymes might contribute to an enforced capacity of differentiated cells to synthesise branched poly-N-acetyllactosamine N-glycans chains. Interestingly, the polysialyltransferase ST8Sia IV (#57), one of the key enzymes of polysialic acid (PSA) biosynthesis, a structure usually found on neural cell adhesion molecule [53], is over-expressed in DCs. However, in contrast with a recent report describing PSA expression on neuropilin-2 [54], we failed to detect PSA expression on the DC surface (not shown). This contrasting result may be explained by differences in the experimental protocols used to generate DCs, which could result in the lack of neuropilin-2 and therefore of PSA. Differentiation is also accompanied to changes in the expression of GTs involved in O-linked glycan synthesis. Compared to monocytes, the gene encoding the polypeptide N-acetylgalactosaminyltransferase 6 (ppGalNAcT6 or GALNT6, #16), is increased in both DCs and Mϕs. In parallel, ppGalNAcT10 (GALNT10, #12) is decreased in both differentiated cells and ppGalNAcT3 (GALNT3, #7) in DCs. This indicates that, along with an enforced N-type glycosylation, the mucin-type O-linked glycosylation might also be modified during DC and Mϕ differentiation. FUT11, the gene encoding the putative α1,3/4-fucosyltransferase 11 (#3) is also increased in both DCs and Mϕs. This might explain the increased expression of sLex in immature DCs (~27 fold compared to monocytes), as indicated by FACS analysis (Table 3), although the expression other enzymes implicated in sLex synthesis (FUT7, ST3Gal III, IV or VI) remains unchanged. Major changes are also found for genes encoding enzymes implicated in proteoglycan synthesis, in particular in the sulfatation of GAGs. Indeed, the HS sulfotransferase HS2ST1 (#70) is increased in both DCs and Mϕs, whilst the chondroitin 4-sulfotransferase CHST12 (#72) is increased only in DCs. Conversely, transcripts for the N-acetylgalactosamine-4-sulfate 6-O-sulfotransferase (GalNAc4S6ST, #80), the N-acetylglucosamine 6-O-sulfotransferase 2 (CHST2, #77) and the chondroitin sulfate N-acetylgalactosaminyltransferase 1 (CHGN, #82) are decreased in differentiated cells, relative to monocytes. Finally, transcript for exostosin-like 2 (EXTL2, #81), a copolymerase that transfers GlcNAc and GlcA to the common GAG-protein linkage region to initiate HS synthesis [55], is increased in both DCs and Mϕs, whilst the heparan sulfate copolymerase EXT1 (#87) is decreased in Mϕs. In parallel, within the proteoglycan family, a group of genes including agrin, bamacan, glypican 4, decorin and syndecan 2, 3 and 4 are upregulated in both DCs and Mϕs, as revealed by our micro-array analysis (not shown). Altogether, in agreement with Wegrowski et al. [56], DCs and Mϕs probably exhibit an important remodelling of proteoglycans on their surface.
Heatmaps showing differential regulation of GT and sulfotransferase gene expression during the differentiation of monocytes into DCs and Mϕs. Monocytes were cultured to differentiate into either DCs (GM-CSF plus IL-4) or Mϕs (GM-CSF). Profiles for transcripts differentially expressed in both DCs and Mϕs (a), only in DCs (b) and only in Mϕs (c). Heatmap colors represent relative log2 expression values median scaled across the samples for each transcript, red showing increased and blue showing decreased expression. Common names and probe #’s from Table 1 are indicated beside the heatmaps. Details for determining these significant differentially transcripts are presented in the Methods section
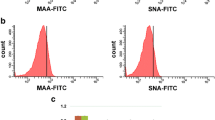
Comparison of gene expression between DCs and Mϕs also indicates that a limited number of GT and sulfotransferase genes are cell specific (12 in DCs and 8 in Mϕs, Fig. 1a and c). In agreement with a recent study reporting a high α2,6-linked sialic acid density in immature DCs [57], ST6Gal I (#63) is over-expressed in DCs. The enhanced (~3-fold) exposure of α2,6-linked sialic acid on differentiated cells, relative to monocytes, was confirmed by flow cytometry using the SNA lectin (Table 3). The lactotriaosylceramide (Lc3) synthase (B3GNT5, #24) and the Forssman synthase (GBGT1, #13), the enzyme that converts the Pk Ag into the Forssman Ag, are significantly increased in DCs, but not in Mϕs. These latter observations suggest potential differences in the synthesis of globo and (neo)lacto series of GSLs between DCs and Mϕs, which could have important consequences on the fine tuning of membrane microdomain organization and cell signalling.
3.3 Comparison of GT and sulfotransferase gene expression in mature DCs and Mϕs, relative to immature cells
As a next step, we compared the expression levels of GT and sulfotransferase transcripts in immature versus mature DCs or Mϕs. To this end, cells were stimulated for 18 h with the canonical TLR4 agonist lipopolysaccharide (LPS). As expected, LPS strongly increases the expression of CD80 and CD86 in DCs, and at a lower level in Mϕs, whilst that of HLA-DR and CD83 was only induced in mature DCs (Table 3). Microarray analysis revealed that the expression of lectins, which are upregulated during the differentiation, is mainly repressed after maturation (not shown), as confirmed by flow cytometry for DC-SIGN (CD209) (Table 3). Thus, LPS treatment results in dramatic changes in DC and Mϕ phenotype.
Compared to immature cells, LPS treatment is accompanied by a variation of some GT and sulfotransferase gene expression, in particular in DCs. Among modulated genes, 23 genes in DCs and 25 genes in Mϕs are significantly decreased after stimulation, respectively (Fig. 2). On the other hand, in DCs and Mϕs, respectively, 15 and 6 genes are increased at 18 h compared to resting cells. Most of these variations were confirmed by qPCR on RNA from three to five samples different from those used in the gene chips analysis (Table 5). Strikingly, LPS treatment preferentially down-modulated genes found to be positively regulated during the differentiation steps (nine for DCs and 11 for Mϕs). As an example, MGAT4A (#27), DDOST (#96), DAD1 (#92), GLCE (#89), HS2ST1 (#69) and B3GNT1 (#29), which are increased during DC and Mϕ differentiation, are decreased in LPS-stimulated DCs and Mϕs. A similar observation is made for ST6Gal I (#63), which is increased during the differentiation steps, but strongly decreased after LPS treatment. This was however not accompanied by a decreased binding of SNA on mature cells (Table 3). With the notable exception of core 1 β3Gal T1 (C1GALT1, #18) and ST3GAL1 (#65), which are increased in stimulated DCs, most of the O-linked glycan enzymes are decreased after stimulation. In particular, the core 2 β6 GlcNAc T1 (GCNT1, #28) is decreased in both mature DCs and Mϕs. Altogether, these observations are in agreement with [25, 26] and indicate that mature cells have a reduced capacity for synthesising mucin-type O-glycans, which might be reduced to sialylated core 1. In agreement with the general picture that maturation inversely modulates the expression of genes affected during the differentiation step, genes such as B4GALT5 (#24), EXT1 (#87), CHST12 (#72) and GALNT10 (#8), found to be down-regulated during the differentiation steps, are generally increased in mature DCs or Mϕs (Fig. 2). However, notable exceptions are observed, for example for genes only modulated during the maturation, but not during the differentiation process. These genes include FUT4 (#6), an α3-fucosyltransferase involved in the synthesis of selectin ligands, and the core 1 β3-Gal T1 (#18), for which expression is unchanged during differentiation, but increased in both mature DCs and Mϕs. We also noticed that some genes including B3GNT2 (#30), the sulfotransferase CHST12 (#72), B4GALT4 (#17), the copolymerase EXTL2 (#81) and ST8SIA4 (#57), found to be enhanced during the differentiation, are also increased during the maturation of DCs and/or Mϕs (Fig. 2). Finally, LPS treatment leads to cell-specific differences. The first difference concerns the expression of genes, including the pp-GalNAc T2 (#11), the β3-GlcNAc T2 (#30), the N-acetylglucosamine 6-O-sulfotransferase 7 (CHST7, #74) and 14 (CHST14, #73), the sialyltransferases ST6GalNAc II (#61) and ST3Gal VI (#58) and the chondroitin GalNAc-transferase 2 (CSGALNACT2, #83), which are strongly modified (positively or negatively) in LPS-treated DCs (Fig. 2b), but unchanged in stimulated Mϕs (Fig. 2c). Similarly, compared to Mϕs, transcripts for some GSL-specific GTs are enhanced in mature DCs, but not Mϕs. These include the glucosylceramide synthase UGCG (#43) and the GM3 synthase (ST3GAL5, #64), the later being involved in the synthesis of ganglio series (a, b and c) of GSLs. Flow cytometry analysis, however, revealed no significant increase of GM3 cell surface expression after LPS stimulation and no GD3 synthesis, in correlation with the low expression of ST8SIA1. Although not fully GSL specific, ST3Gal I (#65), which is implicated in the synthesis of complex gangliosides (GM1b/GD1a/GT1b/GQ1c) is also increased in DCs, but not in Mϕs.
Heatmaps showing differential regulation of GT and sulfotransferase gene expression during the LPS-induced maturation of DCs or Mϕs. Resting DCs or Mϕs were stimulated with LPS for 18 h. Profiles for transcripts differentially expressed in both DCs and Mϕs (a), only in DCs (b) and only in Mϕs (c). For details see legend for Fig. 1. Genes modulated in opposite ways during differentiation and maturation are marked by an asterisk
4 Discussion
Using a highly sensitive array gathering probes for 175 genes involved in the biosynthesis of N-glycans, O-glycans, GAG and glycolipids, we first analyzed GT and sulfotransferase gene expression profile of human monocytes. Microarray analysis indicated that, among the 175 genes examined, 90 are significantly expressed in human monocytes. Monocytes express transcripts for a full set of enzymes involved in the biosynthesis of multiantennary N-glycans, possibly elongated by poly-N-acetyl-lactosamine chains, and of Core 1 and Core 2 O-linked sialylated glycan chains. Monocytes also express genes potentially involved in sLex, but not (s)Lea, Leb and Ley, biosynthesis, a finding confirmed by FACS analysis. The synthesis of sLex on monocytes might be important in the extravasation and infiltration of these cells into peripheral sites, where DCs and Mϕs differentiate [27]. Monocytes also synthesize genes encoding GTs involved in GAG (heparin and chondroitin sulfate) biosynthesis and encoding sulfotransferases involved in their modification. On the other hand, judging by the low expression levels of genes encoding GT specifically implicated in GSL synthesis, monocytes seem to display a limited capacity to produce glycolipids.
Because differentiation of DCs and Mϕs is accompanied by programmed remodelling of cell surface glycans with biologically important consequences, we next compared GT and sulfotransferase gene expression in in vitro differentiated cells, relative to monocytes. Previous reports have shown that monocyte-to-Mϕ, and possibly monocyte-to-DC differentiation, is associated with modulation of ~1 to 2 % of the global transcriptome [58]. We found that, among genes found to be significantly expressed in monocytes, ~30 % are modulated (mostly increased) in both DCs and Mϕs. These gene array data were validated by qPCR using biological samples independent of those used in the gene chips analysis. This suggests that GT and sulfotransferase genes are relatively sensitive to the differentiation steps and that DCs and Mϕs present an enhanced capacity to produce glycans compared to monocytes. Among biosynthetic pathways, it seems that globally, transcripts for enzymes involved in N-glycosylation pathway, as well as in mucin-type O-glycan and GAG synthesis, are mainly affected. In particular, GT genes involved in N-glycan synthesis are generally increased in differentiated cells. The general picture is that transcripts encoding enzymes involved in both initiation and elongation of N- and O-glycan chains are enhanced during differentiation. Genes encoding endoplasmic reticulum enzymes involved in dolichol cycle or encoding oligosaccharidyltransferase subunits are increased in differentiated cells. In the same time, enzymes involved in both branching and elongation of poly-N-acetyllactosamine chains of N-glycans, are also augmented. MALDI-TOF analysis of N-glycans in DCs has previously shown the presence of sialylated tri- and tetra-antennary N-glycans, potentially elongated with poly-N-acetyllactosamine and decorated with Lewis-type epitopes [25]. Similarly, several genes encoding polypeptide N-acetylgalactosaminyltransferases are increased in differentiated cells suggesting an enhanced synthesis of O-glycans. Micro-array analysis also shows the preferential expression of several enzymes involved in type II (Galβ1-4GlcNAc) terminal unit biosynthesis in monocytes derived-cells. Altogether, these data indicate an enforced capacity of differentiated cells to synthesize type II blood group epitopes exposed at the surface that fits well with the highest sLex expression in immature DCs and Mϕs, as indicated by FACS analysis. It is possible that, through selectin member interactions, sLex might be involved in the migration of DCs and Mϕs and/or in their interactions with immune cell types (including T cells) [59]. Interestingly enough, specific enzymes involved in the synthesis of other glycan determinants potentially exposed on the cell surface to interact with counter-receptors are positively modulated in differentiated cells. For instance, over-expression of ST6GAL1 transcript in immature DCs, an enzyme that elaborates the terminal sequence NeuAcα2-6Galβ1-4GlcNAc on N-linked (such as CD45) and O-linked glycoproteins, is in agreement with the high α2-6-linked sialic acid density in immature DCs [57] and could be important in many situations. Indeed, this sialylated sequence represents the glycan ligand for CD22 (Siglec-2), a molecule known to negatively regulate signalling events [15] and implicated in the regulation of Siglec-mediated cell death [60]. Within the proteoglycan family, important changes in the expression of genes encoding enzymes implicated in the sulfatation of GAGs and in the synthesis of proteoglycans (i.e. members of the syndecan family) are observed in both DCs and Mϕs. This might result in important remodelling of proteoglycans on their surface, potentially giving new counter-receptors, relative to monocytes. Collectively, although monocytes, DCs and Mϕs all belong to the mononuclear phagocyte system, they display a distinct profile of GT and sulfotransferase expression as a result of cell differentiation. Micro-array analysis has revealed clear analogies between DCs and Mϕs in terms of GT and sulfotransferase transcript expression, without a real clear gene expression signature. Whether the few differences in gene expression between DCs and Mϕs could account for functional differences is an open question that deserves further investigations.
Since glycosylation-related genes are modulated during stress conditions [61–63], we next investigated whether GT and sulfotransferase transcripts could be modulated in DCs and Mϕs in response to LPS. Maturation/activation of DCs and Mϕs results in a profound remodelling of cell surface markers (chemokine receptors, co-stimulatory molecules) with important functional consequences. However, the impact of maturation/activation on glycan synthesis in these cells has only been recently investigated for DCs [25, 26]. To address this question more in depth, cells were stimulated with LPS, one of the most potent TLR activator. Compared to immature cells, LPS treatment was accompanied by a general decrease of GT and sulfotransferase gene expression in both DCs and Mϕs and mainly affects genes that were found to be modulated during DC and Mϕ differentiation. For instance, in DCs, ~40 % of genes increased during differentiation are decreased during maturation. Notably, excepted GTs involved in the synthesis of sialylated core 1 O-glycans, most enzymes involved in O-linked glycosylation, including the core 2 β6 GlcNAc T1 (GCNT1), are decreased after LPS stimulation. These findings are in line with recent reports [25, 26], although our data do not confirm the total loss of sLex, known to require core 2 structures, on mature DCs. This discrepancy may be explained by experimental differences to induce DC maturation (TNF-α plus PGE2 vs. LPS). Of note, several genes are also increased during DC maturation such as those involved in GAG biosynthesis (sulfotransferases and co-polymerases). This suggests an enhanced synthesis of HS in mature cells that can be implicated in signaling functions of heparin-binding growth factors and chemokines [55, 64]. Transcripts for enzymes implicated in the production of globo-series (B3GALNT1, GBGT1) and ganglio-series of GSLs (ST3GAL5, ST3GAL1) are specifically enhanced in mature DCs, relative to Mϕs. This, and the enhanced expression of ceramide glucosyltransferase (UGCG) in DCs, strongly suggests that a profound change of GSL metabolism operates in DCs undergoing maturation. The relevance of this finding is still unknown and deserves further investigations. Glycolipids have been clearly demonstrated to be involved in differentiation, proliferation and migration of neural cells in mammalian central nervous system, and changes in GSL expression in mature DCs might be important in their immunostimulatory functions. Interestingly, TLR-stimulated DCs have recently been shown to generate new or increasing amounts of glycolipid Ags able to activate different subsets of lipid-reactive innate/memory T cells, via the CD1 molecules [65–69]. This pathway is believed to play an important role in innate responses to pathogens and to polarize acquired responses [23, 70]. Altogether, these data suggest that by affecting certain enzymes of the GSL pathway, TLR triggering in DCs could play an important part in innate/acquired responses.
In conclusion, we herein report the GT and sulfotransferase gene expression profiling in DCs and Mϕs, both in basal and stress (pro-inflammatory) conditions. We show that differentiation (monocyte-to-DC and monocyte-to-Mϕ) and maturation processes are associated with significant changes in GT and sulfotransferase gene expression. Interestingly, although DCs and Mϕs express a comparable pattern of enzymes, cell specific genes are also differentially modulated during the differentiation and maturation processes, a phenomenon that may sustain functional specificities.
Abbreviations
- APC:
-
Antigen presenting cell
- DC:
-
Dendritic cell
- Ag:
-
Antigen
- GSL:
-
Glycosphingolipid
- GT:
-
Glycosyltransferase
- Mϕ:
-
Macrophage
- TLR:
-
Toll-like receptor
- sLex :
-
Sialyl Lex
- GAG:
-
Glycosaminoglycan
- ST:
-
Sialyltransferase
- PSA:
-
Polysialic acid
- HS:
-
Heparan sulfate
- qPCR:
-
Quantitative real-time PCR
- LPS:
-
Lipopolysaccharide
- MFI:
-
Mean fluorescence intensity
- GM-CSF:
-
Granulocyte Mϕ-colony stimulating factor
References
Arnold, J.N., Wormald, M.R., Sim, R.B., Rudd, P.M., Dwek, R.A.: The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25, 21–50 (2007). doi:10.1146/annurev.immunol.25.022106.141702
Collins, B.E., Paulson, J.C.: Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr. Opin. Chem. Biol. 8, 617–625 (2004). doi:10.1016/j.cbpa.2004.10.004
Daniels, M.A., Hogquist, K.A., Jameson, S.C.: Sweet ’n’ sour: the impact of differential glycosylation on T cell responses. Nat. Immunol. 3, 903–910 (2002). doi:10.1038/ni1002-903
Marth, J.D., Grewal, P.K.: Mammalian glycosylation in immunity. Nat. Rev. Immunol. 8, 874–887 (2008). doi:10.1038/nri2417
Rudd, P.M., Elliott, T., Cresswell, P., Wilson, I.A., Dwek, R.A.: Glycosylation and the immune system. Science 291, 2370–2376 (2001). doi:10.1126/science.291.5512.2370
Spiro, R.G.: Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R (2002). doi:10.1093/glycob/12.4.43R
Crocker, P.R.: Siglecs: sialic-acid-binding immunoglobulin-like lectins in cell-cell interactions and signalling. Curr. Opin. Struct. Biol. 12, 609–615 (2002). doi:10.1016/S0959-440X(02)00375-5
Esko, J.D., Selleck, S.B.: Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 (2002). doi:10.1146/annurev.biochem.71.110601.135458
Lau, K.S., Partridge, E.A., Grigorian, A., Silvescu, C.I., Reinhold, V.N., Demetriou, M., Dennis, J.W.: Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 (2007). doi:10.1016/j.cell.2007.01.049
Lowe, J.B.: Glycosyltransferases and glycan structures contributing to the adhesive activities of L-, E- and P-selectin counter-receptors. Biochem. Soc. Symp. 69, 33–45 (2002)
Moody, A.M., Chui, D., Reche, P.A., Priatel, J.J., Marth, J.D., Reinherz, E.L.: Developmentally regulated glycosylation of the CD8alphabeta coreceptor stalk modulates ligand binding. Cell 107, 501–512 (2001). doi:10.1016/S0092-8674(01)00577-3
Rabinovich, G.A., Baum, L.G., Tinari, N., Paganelli, R., Natoli, C., Liu, F.T., Iacobelli, S.: Galectins and their ligands: amplifiers, silencers or tuners of the inflammatory response? Trends Immunol. 23, 313–320 (2002). doi:10.1016/S1471-4906(02)02232-9
Toscano, M.A., Bianco, G.A., Ilarregui, J.M., Croci, D.O., Correale, J., Hernandez, J.D., Zwirner, N.W., Poirier, F., Riley, E.M., Baum, L.G., Rabinovich, G.A.: Differential glycosylation of TH1, TH2 and TH-17 effector cells selectively regulates susceptibility to cell death. Nat. Immunol. 8, 825–834 (2007). doi:10.1038/ni1482
Blander, J.M., Visintin, I., Janeway Jr., C.A., Medzhitov, R.: Alpha(1, 3)-fucosyltransferase VII and alpha(2, 3)-sialyltransferase IV are up-regulated in activated CD4 T cells and maintained after their differentiation into Th1 and migration into inflammatory sites. J. Immunol. 163, 3746–3752 (1999)
Collins, B.E., Blixt, O., Han, S., Duong, B., Li, H., Nathan, J.K., Bovin, N., Paulson, J.C.: High-affinity ligand probes of CD22 overcome the threshold set by cis ligands to allow for binding, endocytosis, and killing of B cells. J. Immunol. 177, 2994–3003 (2006)
Feizi, T.: Carbohydrate-mediated recognition systems in innate immunity. Immunol. Rev. 173, 79–88 (2000). doi:10.1034/j.1600-065X.2000.917310.x
Moody, A.M., North, S.J., Reinhold, B., Van Dyken, S.J., Rogers, M.E., Panico, M., Dell, A., Morris, H.R., Marth, J.D., Reinherz, E.L.: Sialic acid capping of CD8beta core 1-O-glycans controls thymocyte-major histocompatibility complex class I interaction. J. Biol. Chem. 278, 7240–7246 (2003). doi:10.1074/jbc.M210468200
Morgan, R., Gao, G., Pawling, J., Dennis, J.W., Demetriou, M., Li, B.: N-acetylglucosaminyltransferase V (Mgat5)-mediated N-glycosylation negatively regulates Th1 cytokine production by T cells. J. Immunol. 173, 7200–7208 (2004)
Pappu, B.P., Shrikant, P.A.: Alteration of cell surface sialylation regulates antigen-induced naive CD8+ T cell responses. J. Immunol. 173, 275–284 (2004)
van Kooyk, Y., Rabinovich, G.A.: Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 9, 593–601 (2008). doi:10.1038/ni.f.203
Demetriou, M., Granovsky, M., Quaggin, S., Dennis, J.W.: Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409, 733–739 (2001). doi:10.1038/35055582
Moretta, L., Bottino, C., Pende, D., Castriconi, R., Mingari, M.C., Moretta, A.: Surface NK receptors and their ligands on tumor cells. Semin. Immunol. 18, 151–158 (2006). doi:10.1016/j.smim.2006.03.002
Bendelac, A., Savage, P.B., Teyton, L.: The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007). doi:10.1146/annurev.immunol.25.022106.141711
Godfrey, D.I., Kronenberg, M.: Going both ways: immune regulation via CD1d-dependent NKT cells. J. Clin. Invest. 114, 1379–1388 (2004)
Bax, M., Garcia-Vallejo, J.J., Jang-Lee, J., North, S.J., Gilmartin, T.J., Hernandez, G., Crocker, P.R., Leffler, H., Head, S.R., Haslam, S.M., Dell, A., van Kooyk, Y.: Dendritic cell maturation results in pronounced changes in glycan expression affecting recognition by siglecs and galectins. J. Immunol. 179, 8216–8224 (2007)
Julien, S., Grimshaw, M.J., Sutton-Smith, M., Coleman, J., Morris, H.R., Dell, A., Taylor-Papadimitriou, J., Burchell, J.M.: Sialyl-Lewis(x) on P-selectin glycoprotein ligand-1 is regulated during differentiation and maturation of dendritic cells: a mechanism involving the glycosyltransferases C2GnT1 and ST3Gal I. J. Immunol. 179, 5701–5710 (2007)
Gordon, S., Taylor, P.R.: Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 (2005). doi:10.1038/nri1733
Hume, D.A., Ross, I.L., Himes, S.R., Sasmono, R.T., Wells, C.A., Ravasi, T.: The mononuclear phagocyte system revisited. J. Leukoc. Biol. 72, 621–627 (2002)
Randolph, G.J., Beaulieu, S., Lebecque, S., Steinman, R.M., Muller, W.A.: Differentiation of monocytes into dendritic cells in a model of transendothelial trafficking. Science 282, 480–483 (1998). doi:10.1126/science.282.5388.480
Randolph, G.J., Inaba, K., Robbiani, D.F., Steinman, R.M., Muller, W.A.: Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity 11, 753–761 (1999). doi:10.1016/S1074-7613(00)80149-1
Banchereau, J., Steinman, R.M.: Dendritic cells and the control of immunity. Nature 392, 245–252 (1998). doi:10.1038/32588
Kapsenberg, M.L.: Dendritic-cell control of pathogen-driven T-cell polarization. Nat. Rev. Immunol. 3, 984–993 (2003). doi:10.1038/nri1246
Reis e Sousa, C.: Dendritic cells in a mature age. Nat. Rev. Immunol. 6, 476–483 (2006). doi:10.1038/nri1845
Rossi, M., Young, J.W.: Human dendritic cells: potent antigen-presenting cells at the crossroads of innate and adaptive immunity. J. Immunol. 175, 1373–1381 (2005)
Iwasaki, A., Medzhitov, R.: Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987–995 (2004). doi:10.1038/ni1112
Takeda, K., Akira, S.: Toll-like receptors in innate immunity. Int. Immunol. 17, 1–14 (2005). doi:10.1093/intimm/dxh186
Gosset, P., Bureau, F., Angeli, V., Pichavant, M., Faveeuw, C., Tonnel, A.B., Trottein, F.: Prostaglandin D2 affects the maturation of human monocyte-derived dendritic cells: consequence on the polarization of naive Th cells. J. Immunol. 170, 4943–4952 (2003)
Sallusto, F., Lanzavecchia, A.: Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179, 1109–1118 (1994). doi:10.1084/jem.179.4.1109
Young, D.A., Lowe, L.D., Clark, S.C.: Comparison of the effects of IL-3, granulocyte-macrophage colony-stimulating factor, and macrophage colony-stimulating factor in supporting monocyte differentiation in culture. Analysis of macrophage antibody-dependent cellular cytotoxicity. J. Immunol. 145, 607–615 (1990)
Turville, S.G., Cameron, P.U., Handley, A., Lin, G., Pohlmann, S., Doms, R.W., Cunningham, A.L.: Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3, 975–983 (2002). doi:10.1038/ni841
van Kooyk, Y., Geijtenbeek, T.B.: DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3, 697–709 (2003). doi:10.1038/nri1182
Fleetwood, A.J., Lawrence, T., Hamilton, J.A., Cook, A.D.: Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 178, 5245–5252 (2007)
Verreck, F.A., de Boer, T., Langenberg, D.M., Hoeve, M.A., Kramer, M., Vaisberg, E., Kastelein, R., Kolk, A., de Waal-Malefyt, R., Ottenhoff, T.H.: Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco) bacteria. Proc. Natl. Acad. Sci. USA 101, 4560–4565 (2004). doi:10.1073/pnas.0400983101
Lockhart, D.J., Dong, H., Byrne, M.C., Follettie, M.T., Gallo, M.V., Chee, M.S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., Brown, E.L.: Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14, 1675–1680 (1996). doi:10.1038/nbt1296-1675
Hess, A., Iyer, H.: Fisher’s combined p-value for detecting differentially expressed genes using Affymetrix expression arrays. BMC Genomics 8, 96 (2007). doi:10.1186/1471-2164-8-96
Bolstad, B.M., Irizarry, R.A., Astrand, M., Speed, T.P.: A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). doi:10.1093/bioinformatics/19.2.185
Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., Speed, T.P.: Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 (2003). doi:10.1093/nar/gng015
Elbim, C., Hakim, J., Gougerot-Pocidalo, M.A.: Heterogeneity in Lewis-X and sialyl-Lewis-X antigen expression on monocytes in whole blood: relation to stimulus-induced oxidative burst. Am. J. Pathol. 152, 1081–1090 (1998)
Skacel, P.O., Edwards, A.J., Harrison, C.T., Watkins, W.M.: Enzymic control of the expression of the X determinant (CD15) in human myeloid cells during maturation: the regulatory role of 6-sialytransferase. Blood 78, 1452–1460 (1991)
Becker, S., Warren, M.K., Haskill, S.: Colony-stimulating factor-induced monocyte survival and differentiation into macrophages in serum-free cultures. J. Immunol. 139, 3703–3709 (1987)
Inaba, K., Inaba, M., Romani, N., Aya, H., Deguchi, M., Ikehara, S., Muramatsu, S., Steinman, R.M.: Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 176, 1693–1702 (1992). doi:10.1084/jem.176.6.1693
Sasaki, K., Kurata-Miura, K., Ujita, M., Angata, K., Nakagawa, S., Sekine, S., Nishi, T., Fukuda, M.: Expression cloning of cDNA encoding a human beta-1, 3-N-acetylglucosaminyltransferase that is essential for poly-N-acetyllactosamine synthesis. Proc. Natl. Acad. Sci. USA 94, 14294–14299 (1997). doi:10.1073/pnas.94.26.14294
Weinhold, B., Seidenfaden, R., Rockle, I., Muhlenhoff, M., Schertzinger, F., Conzelmann, S., Marth, J.D., Gerardy-Schahn, R., Hildebrandt, H.: Genetic ablation of polysialic acid causes severe neurodevelopmental defects rescued by deletion of the neural cell adhesion molecule. J. Biol. Chem. 280, 42971–42977 (2005). doi:10.1074/jbc.M511097200
Curreli, S., Arany, Z., Gerardy-Schahn, R., Mann, D., Stamatos, N.M.: Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 282, 30346–30356 (2007). doi:10.1074/jbc.M702965200
Kitagawa, H., Shimakawa, H., Sugahara, K.: The tumor suppressor EXT-like gene EXTL2 encodes an alpha1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J. Biol. Chem. 274, 13933–13937 (1999). doi:10.1074/jbc.274.20.13933
Wegrowski, Y., Milard, A.L., Kotlarz, G., Toulmonde, E., Maquart, F.X., Bernard, J.: Cell surface proteoglycan expression during maturation of human monocytes-derived dendritic cells and macrophages. Clin. Exp. Immunol. 144, 485–493 (2006). doi:10.1111/j.1365-2249.2006.03059.x
Jenner, J., Kerst, G., Handgretinger, R., Muller, I.: Increased alpha2, 6-sialylation of surface proteins on tolerogenic, immature dendritic cells and regulatory T cells. Exp. Hematol. 34, 1212–1218 (2006). doi:10.1016/j.exphem.2006.04.016
Martinez, F.O., Gordon, S., Locati, M., Mantovani, A.: Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 (2006)
Suzuki, A., Yamakawa, M., Tsukamoto, M.: The adhesion molecules, l-selectin and sialyl lewis x, relate to the formation of the follicular dendritic cell-lymphocyte cluster in the mantle zone. Immunol. Lett. 79, 181–187 (2001). doi:10.1016/S0165-2478(01)00282-6
Crocker, P.R.: Siglecs in innate immunity. Curr. Opin. Pharmacol. 5, 431–437 (2005). doi:10.1016/j.coph.2005.03.003
Campbell, B.J., Yu, L.G., Rhodes, J.M.: Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconj. J. 18, 851–858 (2001). doi:10.1023/A:1022240107040
Coulouarn, C., Lefebvre, G., Derambure, C., Lequerre, T., Scotte, M., Francois, A., Cellier, D., Daveau, M., Salier, J.P.: Altered gene expression in acute systemic inflammation detected by complete coverage of the human liver transcriptome. Hepatology 39, 353–364 (2004). doi:10.1002/hep.20052
Van Dijk, W., Brinkman-Van der Linden, E.C., Havenaar, E.C.: Occurrence and possible function of inflammation-induced expression of sialyl Lewis-x on acute-phase proteins. Adv. Exp. Med. Biol 435, 145–150 (1998)
Kim, B.T., Kitagawa, H., Tanaka, J., Tamura, J., Sugahara, K.: In vitro heparan sulfate polymerization: crucial roles of core protein moieties of primer substrates in addition to the EXT1-EXT2 interaction. J. Biol. Chem. 278, 41618–41623 (2003). doi:10.1074/jbc.M304831200
Brigl, M., Bry, L., Kent, S.C., Gumperz, J.E., Brenner, M.B.: Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat. Immunol. 4, 1230–1237 (2003). doi:10.1038/ni1002
De Libero, G., Moran, A.P., Gober, H.J., Rossy, E., Shamshiev, A., Chelnokova, O., Mazorra, Z., Vendetti, S., Sacchi, A., Prendergast, M.M., Sansano, S., Tonevitsky, A., Landmann, R., Mori, L.: Bacterial infections promote T cell recognition of self-glycolipids. Immunity 22, 763–772 (2005). doi:10.1016/j.immuni.2005.04.013
Mattner, J., Debord, K.L., Ismail, N., Goff, R.D., Cantu 3rd, C., Zhou, D., Saint-Mezard, P., Wang, V., Gao, Y., Yin, N., Hoebe, K., Schneewind, O., Walker, D., Beutler, B., Teyton, L., Savage, P.B., Bendelac, A.: Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434, 525–529 (2005). doi:10.1038/nature03408
Paget, C., Mallevaey, T., Speak, A.O., Torres, D., Fontaine, J., Sheehan, K.C., Capron, M., Ryffel, B., Faveeuw, C., Leite de Moraes, M., Platt, F., Platt, F., Trottein, F., Trottein, F.: Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity 27, 597–609 (2007). doi:10.1016/j.immuni.2007.08.017
Salio, M., Speak, A.O., Shepherd, D., Polzella, P., Illarionov, P.A., Veerapen, N., Besra, G.S., Platt, F.M., Cerundolo, V.: Modulation of human natural killer T cell ligands on TLR-mediated antigen-presenting cell activation. Proc. Natl. Acad. Sci. USA 104, 20490–20495 (2007). doi:10.1073/pnas.0710145104
Tupin, E., Kinjo, Y., Kronenberg, M.: The unique role of natural killer T cells in the response to microorganisms. Nat. Rev. Microbiol. 5, 405–417 (2007). doi:10.1038/nrmicro1657
Garcia-Vallejo, J.J., Gringhuis, S.I., van Dijk, W., van Die, I.: Gene expression analysis of glycosylation-related genes by real-time polymerase chain reaction. Methods Mol. Biol. 347, 187–209 (2006)
Groux-Degroote, S., Krzewinski-Recchi, M.A., Cazet, A., Vincent, A., Lehoux, S., Lafitte, J.J., Van Seuningen, I., Delannoy, P.: IL-6 and IL-8 increase the expression of glycosyltransferases and sulfotransferases involved in the biosynthesis of sialylated and/or sulfated Lewisx epitopes in the human bronchial mucosa. Biochem. J. 410, 213–223 (2008). doi:10.1042/BJ20070958
Acknowledgments
We acknowledge Dr Juan J. Garcia-Vallejo (VU Medical Center, Amsterdam, The Netherlands) for the gift of some oligonucleotides used for the qPCR analysis. We also thank Pr. R. Gerardy-Schahn (Medizinische Hochschule, Hannover, Germany) for the gift of the anti-PSA Ab.
This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Pasteur Institute of Lille, the University of Lille 2, the Contrat de Plan Etat Région 2000–2006 (CPER)/FEDER (Fonds Européen de Développement Régional) and l’Agence Nationale de la Recherche (ANR) (program MIE grant R08066ES/RPV08036ESA). This work was also supported by Consortium for Functional Glycomics Grant GM-62116 from the National Institutes of Health. CP was recipient of a doctoral fellowship from the Conseil Régional Nord—Pas-de-Calais / INSERM. FT is supported by the Centre National de la Recherche Scientifique, CF and PG by the INSERM and SGD, MAKR and PD by the University of Lille 1.
Conflict of interest statement
None declared.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Trottein, F., Schaffer, L., Ivanov, S. et al. Glycosyltransferase and sulfotransferase gene expression profiles in human monocytes, dendritic cells and macrophages. Glycoconj J 26, 1259–1274 (2009). https://doi.org/10.1007/s10719-009-9244-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-009-9244-y