Abstract
In addition to their roles in developmental and metabolic processes, MYB transcription factors play crucial roles in plant defense mechanisms and stress responses. A comprehensive analysis of six pearl millet genomes revealed the presence of 1133 MYB genes, which can be classified into four phylogenetically distinct subgroups. The duplication pattern of MYB genes across the pearl millet genomes demonstrates their conserved and similar evolutionary history. Overall, MYB genes were observed to be involved in drought and heat stress responses, with stronger differential expressed observed in root tissues. Multiple analyses indicated that MYB genes mediate abiotic stress responses by modulating abscisic acid-related pathways, circadian rhythms, and histone modification processes. A substantial number of duplicated genes were determined to exhibit differential expression under abiotic stress. The consistent positive expression trend observed in duplicated gene pairs, such as PMA5G04432.1 and PMA2G00728.1, across various abiotic stresses suggests that duplicated MYB genes plays a key role in the evolution of adaptive responses of pearl millet to abiotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MYB proteins are characterized as having a DNA-binding domain and are one of the largest families of transcription factors in plants (Karin 1990; Stracke et al. 2001). MYB proteins can be classified into four types based on the number of recognition helices: 1R-MYB, R2R3MYB, 3R-MYB and 4R-MYB (Ogata et al. 1994; Jia et al. 2004; Farhat et al. 2021; Dubos et al. 2010). Additionally, the presence of 5R-MYB has also been investigated (Chai et al. 2020). However, the biological information of 3R-MYB, 4R-MYB and 5R-MYB are still largely unknown. Recently, the increasing availability of plant genome sequences has facilitated a better understanding of this large gene family. In addition to Arabidopsis, genome wide characterization of the MYB family, especially for MYB, has done in rice (Oryza sativa) (Katiyar et al. 2012), barely (Hordeum vulgare) (Tombuloglu et al. 2013), maize (Zea mays) (Du et al. 2012), foxtail millet (Setaria italica) (Muthamilarasan et al. 2017), soybean (Glycine max) (Du et al. 2012), Mango (Mangifera indica) (He et al. 2022) and Kenaf (Hibiscus cannabinus) (Li et al. 2022). Comparative expression profile analysis of MYB genes in these species suggested that MYB proteins play conserved and various roles in development, growth and regulation of the metabolism of plants (Dubos et al. 2010). MYB proteins play indispensable roles in phytohormone signal transduction and various stress response pathways (Ambawat et al. 2013). Most of the current studies found 1R-MYB, 2R-MYB associated with abiotic stresses, for example, R2R3-TaMYB30 was found in wheat through interaction with other genes and R2R3-MdMYB30 was found to play an important role in salt stress response in apple (Wang et al. 2018; Su et al. 2018); overexpression of 1R-MYB1 enhances resistance to salt stress in Arabidopsis thaliana. Transcription factor 1R-MYB15 is involved in the response process of Arabidopsis to drought stress. 1R-MYB15 overexpression enhances Arabidopsis resistance to drought stress and can promote plant growth (Huang et al. 2020; Wei et al. 2016).
Pearl millet (Pennisetum glaucum) is the sixth most important cereal crop in the world after rice, wheat, maize, barley and sorghum (Sehgal et al. 2012; Sun et al. 2020). It is grown as a staple crop in the hottest and driest regions of sub-Saharan Africa and the Indian subcontinent (Dan et al. 2020; Oosterom et al. 1996), where rainfall is very limited (300–500 mm generally) and other crops, such as maize or sorghum, are unlikely to survive (Khan et al. 2023; Mahalakshmi et al. 1987). Natural and artificial selection have enabled pearl millet to become the most tolerant to drought and high temperatures of all food crops (Awan et al. 2022; Merga 2020), making it an ideal plant material for studying abiotic stress responses in cereal crops.
Although studies have identified the MYB gene family in pearl millet and the expression levels of some MYB genes under milder abiotic stresses (Varshney Rajeev et al. 2017; Lin et al. 2023), these analyses were performed using individual second-generation sequenced genomes (Jeky et al. 2023). However, several important questions regarding the evolutionary pattern, tissue expression specificity, and abiotic stress expression characteristics of pearl millet MYB genes remain largely unclear.
At present, based on three-generation sequencing, 10 pearl millet genomic data has been available (Yan et al. 2023), which provides new resources for comprehensive analysis of pearl millet MYB gene families and to investigate their functions in abiotic stresses on a genome-wide scale. In this study we selected three wild types and three cultivated types of pearl millet for a comprehensive analysis of their MYB gene families. The objectives of this study were (1) to investigate the structure, location, and duplication of the six pearl millet MYB members, (2) to investigate their expression patterns and tissue specificity under different levels of abiotic stress.
Material method
Pearl millet MYB gene identification
Genomes and annotation files for six species of pearl millet (Pennisetum glaucum) were downloaded from the Milletdb database (http://117.78.45.2:91/home). The hidden Markov model (HMM) of the MYB family (PF 00249) was obtained from the Pfam database (http://pfam.xfam.org/), and the original MYB HMM was used to obtain protein ensembles (E-values < 1 × 10–20) using the HMMER software tool (Finn et al. 2011; Letunic et al. 2021). The pearl-millet-specific HMMs were constructed using hmmbuild in the HMMER suite based on the MYB HMM. Proteins with E-values below 0.01 were identified using the pearl-millet-specific HMM. The presence of each gene containing the MYB structural domain was confirmed using SMART (http://smart.emblheidelberg.de/) and the Pfam database (Jaina et al. 2021). The isoelectric point and molecular weight of MYB proteins were predicted using the ExPASy server (http://web.expasy.org/compute_pi/).
Multiple sequence alignment and phylogenetic evolution
The R2R3-MYB multiple sequence alignment of sequences from pearl millet was performed with default parameters (bootstrap value, 1000) using clustalX2 software. The phylogenetic tree of pearl millet MYB genes was constructed using the neighbor-joining (NJ) method as implemented in MEGA11 and visualized using iTOL (https://itol.embl.de/itol.cgi) (Letunic and Bor 2021).
Exon/intron structure, structural domain and chromosome localization analysis
The structural information for pearl millet MYB genes was obtained by extracting GFF file gene structure and location information with TBtools (Chen et al. 2020), and the conserved motifs in proteins were identified by MEME (http://MEME-suite.org/meme/) with the motif count set to 20 and other parameters set to default values (Bailey et al. 2009). Finally, the results were visualized using CFVisual software (Chen et al. 2022). Gene chromosome localization was visualized using TBtools.
Species development evolutionary tree, MYB gene duplication, covariance analysis and Ka/Ks analysis
Genomic, proteomic and annotation files of rice, maize, Arabidopsis and wheat were downloaded from the Ensembl database (Yates et al. 2022), while those for purple elephant grass (Cenchrus purpureus) were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov). Phylogenetic trees of MYB orthologs were constructed using the OrthoFinder program (Emms and Kelly 2019), and co-linear relationships of MYB gene locations between species were analyzed using MCScanX with an E-value of 1e−10. The reliability of the co-linear relationships was then verified using the gene duplication results obtained from OrthoFinder (Yupeng et al. 2012), and the co-linear relationships were visualized using TBtools and Circos. Ka and Ks values were calculated using the ParaAT2.0 program (Zhang et al. 2012). MAFFT software was used for sequence alignment (Katoh et al. 2005). KaKs_Calculator was used to calculate the Ka/Ks ratios for duplicate gene pairs (Wang et al. 2010).
Transcriptomic analysis
RNA-seq data obtained from pearl millet under different abiotic stresses (salt, heat, drought) for different time durations (5 h and 96 h) and in different tissues (leaf and root) were downloaded from the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra/). Transcripts per million (TPM) values were obtained from the RNA-seq data, and log2 (fold change) values of genes were calculated using DEseq2 (Love et al. 2014). Finally, a heat map was generated using the pheatmap package in the R statistical computing environment. Functional networks and gene connectivity data for differentially expressed genes in Pennisetum glaucum were extracted from the STRING database (https://cn.string-db.org/), species selected foxtail millet, which provides gene connectivity data based on multiple evidence types (direct interaction, co-localization, gene-regulation and co-citation), with a confidence level of 0.7. These data were then analyzed using the iGraph R package, and network analysis employing edge-betweenness and random walk methods was used to identify subnetworks or neighborhoods. The resulting neighborhoods were tested for enrichment of relevant biological/pathway terms using the STRING database.
Weighted gene co-expression network analysis
Using the same pearl millet RNA-seq data obtained from the NCBI SRA database, fragments per kilobase per million (FPKM) values were calculated using the R language DEseq2 package to select pearl millet MYB gene sample data for subsequent analysis. Co-expression networks were constructed using the WGCNA R package, and hierarchical clustering trees were constructed based on the correlation coefficients between genes by selecting appropriate weighting coefficients β (with a soft threshold) and using a scale-free network distribution. Then, genes with similar expression patterns were grouped into different modules based on the weighted correlation coefficients of the genes. Finally, a topological overlap matrix (TOM) was constructed by converting the similarity matrix into an adjacency matrix using the weighted correlation coefficients. A heat map of the module-sample correlation coefficients was visualized using Pheatmap. Additionally, connectivity was defined as the sum of the weights across all the edges of nodes, and the co-expression network was built using Cytoscape software.
Results
Identification and molecular characterization of the MYB gene family of pearl millet
MYB genes were identified in all six pearl millet genomes. The pearl millet genome was searched and queried using the MYB HMM profile (Pfam:00249) and analyzed using the Pfam and SMART databases to verify the presence of MYB protein structural domains. A total of 1133 MYB proteins were identified (Table S1), and five MYB types were identified based on the number of MYB DNA-binding domains, including 1R-MYB, R2R3-MYB, 3R-MYB, 4RMYB and a few 5RMYB genes (Table 1). Millet variety PI587025 had the fewest MYB proteins, 17 fewer than that of PI526529. A comparison of intron number and protein length revealed that 1RMYB and R2R3MYB are differentiated by only small differences, while 3RMYB and 4RMYB are also similar; in contrast, 5RMYB genes were observed to have the most complex and longest structures.
Phylogenetic relationships of MYB gene family members in pearl millet
To determine the evolutionary relationships of MYB genes in the six genomes, phylogenetic trees were constructed using the neighbor-joining method. All of the MYB gene members identified in PI537069 could be divided into four groups: 1RMYB, R2R3MYB_a, R2R3MYB_b, 3RMYB (Fig. 1A). Similar results were found in the other five pearl millet genomes, while we found 4RMYB and 5RMYB do not form a separate subclade (Fig. S1). In addition, a comprehensive phylogenetic tree constructed based on the MYB members of six pearl millet species was similar to the results of PI537069 (Fig. 1B). As was observed across all six genomes, most of the MYB members in pearl millet were determined to belong to the R2R3MYB subfamily, followed by 1RMYB, 3RMYB, 4RMYB and 5RMYB. Notably, the clustering results show that 3RMYB was more closely related to the R2R3MYB subfamily. 5RMYB and 1RMYB had a closer evolutionary relationship. In line PI537069, 4RMYB is evolutionarily more closely related to 1RMYB, while in other lines it is more closely related to R2R3MYB.
Phylogenetic tree of MYB members in the reference genome PI537069 (A) and six pearl millet (B) genomes. Blue circles indicate 1RMYB gene, red circles indicate R2R3MYB gene, green circles indicate 3RMYB gene and cyan circles indicate 4RMYB gene. Phylogenetic trees were constructed using MEGA11 and the neighbor-joining (NJ) method with 1000 bootstrap replicates
Conserved motifs, structural domains and intron number
To characterize and explore the MYB genes, we analyzed the exon/intron structure of the reference genome PI537069 (Fig. 2). The intron structures were relatively similar within subgroups, with members of the R2R3MYB subgroup having a relatively conserved sequence length and number of introns, and 3RMYB and 1RMYB members having a more complex gene structure. Motif analysis showed that the R2R3MYB motifs are highly conserved, with almost all R2R3MYB genes containing motifs 1, 2 and 3. The 1RMYB subgroup members were found to contain some subgroup-specific motifs, i.e., 11, 12 and 13, which may be associated with the functional divergence of MYB genes. We also found that R2R3MYB and 3RMYB have a more similar motif structure, suggesting that R2R3 and 3RMYB are more closely related.
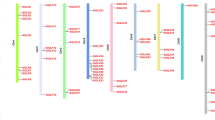
Chromosome distribution analysis
The positions of the identified MYB members were mapped to chromosomes using TBtools, and the MYB gene members were found to be evenly distributed among the seven chromosomes (Fig. 3). Notably, the MYB members occurred in clusters on the chromosomes, suggesting the MYB gene may undergo segmental or tandem duplications. The chromosomal distribution of the MYB gene family within the 6 pearl millet genomes indicate well conservation of MYB family members among the lengthy evolutionary and biological engineering process.
Gene duplication events, syntenic analysis and expansion patterns of the pearl millet MYB genes
To explore the expansion pattern of MYB genes, we analyzed six genomes and found that MYB gene duplication events were mainly clustered on chromosomes 1, 6, and 3. This result was consistent among all six genomes (Fig. 4A). We only observe at least 34 repeats in PI583800, while the rest of the genome exhibits similar duplication events (Fig. 4B). Furthermore, the Ka/Ks ratios among the different genomes were all significantly less than 1 (Fig. 4C, Table S2). Previous research has shown that Ka/Ks ratios below 1 indicate that a gene is under purifying selection (Hurst 2002). Based on these findings, we suggest that pearl millet MYB genes generally experienced purifying selection during their evolution.
To gain a better understanding of the evolutionary relationships of MYB members in different plant species, we compared the reference genome, PI537069, with five genomes of other species (Arabidopsis thaliana, Oryza sativa, Triticum aestivum, Zea mays, Cenchrus purpureus), constructed phylogenetic trees (Fig. 5A) and then compared the collinearity of regions of synteny of each species with Pennisetum glaucum. Pennisetum glaucum was found to have 477 direct homologous pairs with Triticum aestivum (Table S3), which may be related to the genome duplication associated with the hexaploidy of Triticum aestivum, 175 direct homologous pairs with Oryza sativa, 269 direct homologous pairs with Zea mays, 504 direct homologous pairs with Cenchrus purpureus, but only 26 direct homologous pairs with Arabidopsis thaliana, likely owing to the great evolutionary distance separating monocot and dicot plants (Fig. 5B). Additionally, we found a similar number of MYB homologs shared between pearl millet and other graminaceous crops, suggesting that similar evolutionary processes have shaped MYB genes in graminaceous crops. Previous studies have shown that the A subgenome of Cenchrus purpureus is highly homologous to the Pennisetum glaucum genome (Zhang et al. 2022). In the present study we found that Pennisetum glaucum and Cenchrus purpureus were most closely related based on the phylogenetic tree, and based on the genomic co-linearity analysis, 87.85% of all MYB genes were co-linear between the two species, with a high degree of homology, in agreement with previous studies.
Species tree and MYB gene co-linearity comparison. A Species tree of Arabidopsis thaliana, Oryza sativa, Triticum aestivum, Zea mays, Pennisetum glaucum (PI537069) and Cenchrus purpureus, with the number of MYB genes for each species indicated in parentheses. B Co-linearity of MYB genes. The red lines indicate orthologous MYB gene pairs with co-linear relationships. The number beside Pennisetum glaucum indicates the number of MYB genes, while the number beside the other species names indicates the number of co-linear MYB gene pairs
Expression profiles of MYB genes in pearl millet under abiotic stress treatment
To analyze the expression pattern of the pearl millet MYB genes under different abiotic stresses (salt, heat and drought stress) as comprehensively as possible, the expression of PI537069 MYB genes in leaves and roots under different abiotic stress treatment times were compared (after 5 and 96 h of abiotic stress). The transcripts per million (TPM) values were calculated to obtain normalized RNA-seq data (Table S4), and log2(fold change) values (Table S4) were calculated using DESeq2 for roots and leaves at different treatment times to obtain significantly differentially expressed MYB genes; these data are visualized in the form of heat maps in Fig. 6A, C. There were 13 significantly differentially expressed MYB genes after 5 h of abiotic stress, of which 12 (92.31%) were specifically expressed under short-term heat stress. In contrast, only one (7.69%) gene was specifically expressed under short-term drought stress, and no MYB genes were specifically expressed under short-term salt stress (Fig. 6B). MYB genes were more sensitive to mild short-term heat stress and showed an overall down-regulation (83.33%) in leaves and an overall up-regulation (57.14%) in roots. In contrast, under 96-h abiotic stress treatments, 45 MYB genes were significantly differentially expressed, of which 22 (48.89%) were specifically expressed under long-term drought stress, five (11.11%) under long-term heat stress and zero under long-term salt stress (Fig. 6C). We found that six genes (PMA3G07113.1, PMA3G01824.1, PMA4G03528.1, PMA1G06041.1, PMA1G04831.1, PMA1G06161.1) were differentially expressed under each stress treatment, among which four genes (PMA4G03528.1, PMA1G06041.1, PMA1G04831.1, PMA1G06161.1) were differentially expressed in leaf tissue, with PMA4G03528.1 being down-regulated under both drought and heat stresses and only up-regulated under salt stress. Thus, MYB genes exhibited more drastic expression changes under drought stress followed by heat stress and finally salt stress, indicating the relative sensitivity of MYB genes to these three abiotic stresses. These results also suggest that MYB genes have exposure-specific expression patterns.
Differential expression of pearl millet MYB genes under abiotic stress. A Five-hour abiotic stress treatment gene expression heat map (*, |log(fold change)|> 1). B Upset plot displaying the number of differentially expressed MYB genes after the 5-h stress treatment. C Ninety-six-hour stress treatment differential MYB gene expression heat map. D Upset plot displaying the number of differentially expressed MYB genes after the 96-h stress treatment. (Leaf-specific differentially expressed MYB genes are shown in red)
We also conducted a functional annotation-based bioinformatics analysis of the differentially expressed MYB genes using the STRING database. Enrichment analysis revealed that MYB genes differentially expressed under mild abiotic stress mainly affected gene expression and transcription, plant defense, optical morphological changes and circadian pathways (Fig. 7A), indicating that MYB genes may be involved in some key aspects of morphological change and heat stress responses. In response to severe abiotic stress, MYB genes were mainly enriched in modification pathways concerning histone deubiquitination, histone acetylation and DNA methylation (Fig. 7B). Previous research has demonstrated that the induction of stress-responsive genes in plants subjected to abiotic stresses is usually accompanied by changes in the levels of epistatic modifications, particularly histone modifications and DNA methylation levels (Chang et al. 2020). Thus, MYB genes are likely to play an important role in histone modifications in response to abiotic stresses.
Predicted reciprocal network map of differentially expressed MYB genes in pearl millet. A The interaction network based on differentially expressed MYB genes under a 5-h abiotic stress treatment (confidence level = 0.7). Sub-networks (neighborhoods) are colored-coded and annotated for enriched functional categories. B The interaction network based on differentially expressed MYB genes under a 96-h abiotic stress treatment
Weighted gene co-expression network analysis
To further investigate the gene association patterns of pearl millet MYB genes under abiotic stress, a matrix consisting of 36 samples and PI537069 MYB genes was obtained from the NCBI SRA database, and weighted gene co-expression network analysis (WGCNA) was conducted to identify co-expression networks. Normalized data obtained by calculating fragments per kilobase of transcript per million mapped reads (FPKM) values were used as input data for constructing co-expression networks (Table S5). A hierarchical clustering tree of pearl millet consisting of 181 genes from 36 tissue samples was generated (Figure S2). To capture the data in a scale-free way, a β value of 5 was chosen for co-expression network construction. A total of eight modules were obtained by dynamic tree cutting module identification (Figure S3), and genes that could be assigned to any module were placed in the gray module, which was not included in the subsequent statistical analysis. A TOM heat map (Figure S4) was constructed to visualize the interactions among modules. This heat map shows that each module could be identified independently from among the others, indicating their high degree of discreteness and the relative independence of gene expression within each module. In addition, eigengenes were also calculated and clustered based on their correlated expression to explore co-expression similarities between modules; this revealed three clusters were mainly formed by these seven modules (Fig. 8A). A similar result was observed for the adjacency-based heat map (Fig. 8B).
Pearl millet MYB gene co-expression network analysis. A Eigengene neighbor-joining diagram. B Sample module associations, with correlations between samples indicated in red. C Module network diagram of die MYB genes. Node genes are shown in green, while both high and low correlations between them are shown as red lines
As shown in Fig. 8C, there were 9 genes in the green module, 23 in the turquois model (associated with the abiotic stress response in roots) and 5 genes in the red module (associated with the salt-stress response in leaves). Additional modules are shown in Fig. S5, including 4 genes in the black module (associated with leaves and roots during flowering), 13 genes in the yellow module (associated with spikes during flowering), genes in the brown module (associated with spikes during tasseling), 21 genes in the blue module (associated with the development of plants after imbibition). Thus, MYB genes were specific to different stages and tissues, and multiple genes were involved in the co-expression network of root tissues, suggesting that root tissues regulate particularly complex biological processes in responses to abiotic stresses.
Abiotic stress candidate genes
The duplicated MYB gene pair PMA5G04432.1 and PMA2G00728.1 was observed to be differentially expressed in root tissue under heat stress. Thus, we also compared the expression of these genes in the response to heat stress in roots at different times, and both genes had similar expression trends under heat stress treatment (Fig. 9). The OrthoFinder annotation of pearl millet homologous genes in Arabidopsis revealed that the gene pair of PMA5G04432.1 and PMA2G00728.1 is jointly homologous to AT5G67300 (AtMYB44), which has been shown to be involved in drought resistance in Arabidopsis through regulating histone acetylation (Zhao et al. 2022). In the present study, we found this gene pair to be more sensitive to heat stress (Fig. 9), which indicates that these genes may play a more important role in the response to heat stress. Another gene pair, PMA5G00310.1 and PMA3G01824.1, were found to respond to drought stress, but they shared different Arabidopsis homologous genes, AT3G24310 and AT4G13480, respectively (Cheng et al. 2022). Previous research has shown that both genes are upregulated by abscisic acid (ABA) and have high sequence similarity. We found this homologous gene pair was upregulated under multiple stresses, and the expression trends of both were similar (Fig. 9), presumably related to their involvement in regulating ABA sensitivity to resist abiotic stresses. The PMA4G03528.1 homologous gene AT2G16720 is involved in suppressing seed germination in response to ABA and salt stress. We found that this gene also responded to salt stress in pearl millet, but, notably, the expression of this gene was relatively low under other stresses (Fig. 9); thus, the specific stress-response function of this gene needs to be further investigated.
We also counted the number of differentially expressed MYB genes that are duplicates, and 26 duplicate gene pairs (59.09%) were identified. Based on the annotation of their homologous genes in Arabidopsis and a review of previous studies (Zhao et al. 2022; Cheng et al. 2022; Liu et al. 2023; Hamaguchi et al 2008; Mengiste et al. 2003; Lee et al. 2020; Bencan et al. 2018; Wang et al. 2020; Kim et al. 2015; 2020; Perkins et al. 2020; Kim et al. 2015; Qing et al. 2018; Hemm et al. 2001; Xiangbo et al. 2007; Yuhui et al. 2020; Zihang, et al. 2021; Agarwal et al. 2020; Amy et al. 2022), the functions of these pairs were briefly annotated. Thus, 13 duplicate gene pairs (30.95%) are linked to ABA-related pathways, 11 duplicate gene pairs (26.19%) are linked to stress responses, 5 duplicate gene pairs (11.90%) are linked to lignin synthesis, and 4 duplicate gene pairs (9.52%) are linked to plant light response and circadian rhythms (Table S6). These results indicate a that duplicate genes are often involved in responses to abiotic stress and suggest that ABA and MYB genes often function synergistically under abiotic stresses.
Discussion
Identification and phylogenetic analysis of MYB genes in six pearl millet varieties
By constructing a phylogenetic tree of MYB genes in the pearl millet pan-genome, we found the similar structures 1RMYB, R2R3MYB, and 3RMYB subfamilies were identified in different lines, indicating that these three types of genes are conserved in pearl millet. Secondly, structural analysis of MYB genes in pearl millet showed that the same type of MYB genes have similar exon/intron structures. R2R3MYB has the most conserved number of introns, followed by 1RMYB and 3RMYB. 4RMYB and 5RMYB genes have the highest number of introns and complex gene structures. The introns late hypothesis suggests that changes in gene structure are often the result of an expansion in the number of introns (Rogozin et al. 2012; Rosa et al. 2008). In the present study, we note that 3R-MYB formed a separate specific small subfamily, and the difference between 3R-MYB and R2R3-MYB in terms of motif structure mainly involved motif 5, suggesting that the two are evolutionarily more closely related. However, their intron differences are highly substantial, suggesting that 3R-MYB experienced intron gain, likely in association with its divergence from 2R-MYB. Although 4RMYB and 5RMYB do not form a specific subclade, their complex gene structures suggest that they may also be affected by intron gain.
Pearl millet MYB gene evolution and gene duplications in response to abiotic stresses
In addition to polyploidization, gene duplication events are an important engine of evolution (Cannon et al. 2004; Xiong et al. 2008). Our study showed that the number of duplications differed among varieties of pearl millet, with slightly fewer observed in cultivated varieties relative to wild varieties, suggesting that some non-functional MYB genes were lost during domestication. Additionally, duplication was clustered with respect to chromosomal location. Comparison of the number of MYB members in the six pearl millet lines revealed MYB gene family size changes mostly occurred among 1RMYB and R2R3MYB subgroups. The transcriptome analysis showed that genes of these two subgroups were also more associated with responses to abiotic stress, while no 3R-MYB or 4R-MYB members were found to be significantly differentially expressed under abiotic stress. Collectively, these findings suggest that 1R-MYB and R2R3-M MYB duplications may have played a greater role in enhancing abiotic stress responses in pearl millet. We also observed that the number of replication events and replication sites were very similar among the six lines, but produced differences in the number of 1R-MYB and R2R3-MYB genes. However, more in-depth analysis is needed to determine whether this gene family expansion is more associated with natural selection or artificial selection during domestication. Moreover, we identified that many MYB genes exhibited differential expression under abiotic stress, and the gene duplicates were mainly involved in ABA-related, stress defense, and circadian rhythm regulation pathways. Thus, it is hypothesized that such gene duplications maintain the stability of pearl millet growth and development under abiotic stress, which may be critical to the strong abiotic stress resistance of pearl millet.
Expression pattern and tissue specificity of MYB genes under abiotic stress
Previous studies have shown that MYB genes have stage-specific expression and that different genes are expressed differentially under salt stress over time (Yujiao et al. 2021). In the current study we found similar results; the number of genes differentially expressed under intense stress was significantly higher than that under mild stress, and we also found that MYB genes specifically responded to various types of stress. This pattern was not only reflected in the number of genes expressed but also in the specific genes expressed, which changed most under drought and heat stresses, followed by salt stress. In previous research on tea trees, it was found that more MYB genes are expressed in root tissues compared to leaves and stems of normally developing tea trees (Penghui et al. 2022). Our results also showed that MYB genes showed greater specificity of expression in root tissues than leaves, suggesting that MYB genes regulate more complex processes in the roots. For the gene PMA3G07113.1 (AT1G01060.1), which was previously found to influence the microbial environment of the root by regulating circadian rhythm changes in Arabidopsis (Beale et al. 2012), we identified differential expression under each of the three abiotic stresses. Under abiotic stress, both internal conditions of plants as well as external factors can be part of adaptive responses, and we hypothesize that this gene responds to the change in circadian rhythm by regulating interactions with root microbes to maintain plant stability under stress. Notably, we also identified this gene to be differentially expressed in heat-stressed leaves, meriting deeper investigation in further studies.
WGCNA revealed the stage-specific expression of pearl millet not only under abiotic stress, but also during flowering, tasseling and absorption period, which exhibited tissue- and stage-specific expression of MYB genes. For example, PMA3G01387.1 (AT3G60460) was previously found to regulate pollen fertilization in Arabidopsis (Jun et al. 2010), while deletion of PMA4G02292.1 (AT1G06280) in Arabidopsis leads to loss of critical cell wall modifications during flower anther development (Fasani et al. 2019) indicates the functional specificity of MYB genes also exists in plant growth and development.
Conclusions
In this study, a total of 1133 MYB genes were identified from six pearl millet genomes and classified into four subfamilies. The construction of a phylogenetic tree and co-linearity analysis of MYB genes revealed that the pearl millet MYB genes have experienced a highly conserved and similar evolutionary history among graminaceous crops. Subsequently, RNA-seq and WGCNA of pearl millet MYB genes under drought, salt and heat stresses at different time points and in different tissues revealed higher specificity observed in response to drought stress and heat stress and stage-specific expression in different tissues and during developmental stages. Protein interaction prediction and homologous gene annotation analysis indicated that MYB genes are primarily involved in regulating histone modifications that adjust circadian rhythm and ABA-related pathways in response to abiotic stresses. These results reveal key details about the response mechanism of MYB genes under abiotic stress, providing valuable insights into the molecular mechanisms of abiotic stress responses in pearl millet and other gramineous crops.
Data availability
The current dataset for the study can be found in the NCBI database and the Milletdb database, which are publicly accessible, under the following Accession Numbers: PRJNA792845, PRJNA766308, PRJNA756390, PRJNA689619, PRJNA688001.
References
Agarwal P et al (2020) MYB4 transcription factor, a member of R2R3-subfamily of MYB domain protein, regulates cadmium tolerance via enhanced protection against oxidative damage and increases expression of PCS1 and MT1C in Arabidopsis. Plant Sci 297(prepublish):110501
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-013-0179-1
Amy N et al (2022) Circadian rhythms in the plant host influence rhythmicity of rhizosphere microbiota. BMC Biol 20(1):235–235
Awan SA et al (2022) Genome-wide expression and physiological profiling of pearl millet genotype reveal the biological pathways and various gene clusters underlying salt resistance. Front Plant Sci 13:849618
Bailey TL et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(Web Server issue):W202–W208
Beale KM, Leydon AR, Johnson MA (2012) Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Curr Biol 22(12):1090–1094
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4(1):1–21
Chai C, Wang Y, Joshi T, Valliyodan B, Prince S, Michel L et al (2020) Soybean 5R-MYB transcription factors control phosphate acquisition by regulating root architecture and phosphate homeostasis-related gene expression. Plant Cell Environ 43(5):1309–1322. https://doi.org/10.1111/pce.13747
Chang Y-N, Zhu C, Jiang J, Zhang H, Zhu J-K, Duan C-G (2020) Epigenetic regulation in plant abiotic stress responses. J Integrative Plant Biol 62(05):563–580
Chanwala J et al (2023) MYB transcription factor family in pearl millet: genome-wide identification, evolutionary progression and expression analysis under abiotic stress and phytohormone treatments. Plants 12(2):355–355
Chen C et al (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Chen H et al (2022) CFVisual: an interactive desktop platform for drawing gene structure and protein architecture. BMC Bioinform 23(1):178
Cheng Y et al (2022) The R2R3 MYB transcription factor MYB71 regulates abscisic acid response in Arabidopsis. Plants 11(10):1369–1369
Dan X et al (2020) Evaluation of genetic integrity of pearl millet seeds during aging by genomic-SSR markers. Mol Biol Rep 47(8):5747–5754
Du H, Feng B, Yang S, Huang Y, Tang Y (2012) The R2R3-MYB transcription factor gene family in maize. PLoS ONE 7(6):e37463. https://doi.org/10.1371/journal.pone.0037463
Du H, Yang S, Liang Z, Feng B, Liu L, Huang Y, Tang Y (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12(1):106
Dubos C et al (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20(1):238
Fang Q et al (2018) AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling. Plant Cell Rep 37(11):1499–1511
Farhat A et al (2021) Genome-wide analysis reveals the potential role of myb transcription factors in floral scent formation in Hedychium coronarium. Front Plant Sci 12:623742–623742
Fasani E et al (2019) The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol Biol 99(6):517–534
Finn RD, Jody C, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39(Web Server issue):W29-37
Hamaguchi A et al (2008) A small subfamily of Arabidopsis RADIALIS-LIKE SANT/MYB genes: a link to HOOKLESS1-mediated signal transduction during early morphogenesis. Bioscience Biotechnol Biochem. 72(10):2687–2696
Hemm MR, Herrmann KM, Chapple C (2001) AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci 6(4):135–136
Huang J, Zhao X, Cheng K et al (2020) The transcription factor MYB15 is essential for enhancing drought tolerance in Arabidopsis. J Exp Bot 71(6):2087–2102. https://doi.org/10.1093/jxb/eraa041
Hurst LD (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 18(9):486
Jun Z et al (2010) AtMYB103 is a crucial regulator of several pathways affecting Arabidopsis anther development. Sci China Life Sci 53(9):1112–1122
Karin M (1990) Too many transcription factors: positive and negative interactions. New Biologist 2(2):126–131
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom 13(1):544. https://doi.org/10.1186/1471-2164-13-544
Katoh K et al (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acid Res 33(2):511–518
Khan I et al (2023) Physiological and transcriptome analyses demonstrate the silver nanoparticles mediated alleviation of salt stress in pearl millet (Pennisetum glaucum L). Environ Pollut 318:120863–120863
Kim JH, Hyun WY, Nguyen HN et al (2015) AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. https://doi.org/10.1111/pce.12415
Kim SH, Lam PY, Lee MH et al (2020) The Arabidopsis R2R3 MYB transcription factor MYB15 Is a key regulator of lignin biosynthesis in effector-triggered immunity. Front Plant Sci 11:583153
Lee Won WJ et al (2020) NITROGEN RESPONSE DEFICIENCY 1 - mediated CHL1 induction contributes to optimized growth performance during altered nitrate availability in Arabidopsis. Plant J 104(5):1382–1398
Letunic I, Bor P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(W1):W293–W296
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49(D1):D458–D460
Li J, Clegg Michael T, Tao J (2004) Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol 134(2):575–585
Li Hu et al (2022) Transcriptomes of different tissues for expression characteristics analysis of myb gene family in Kenaf (Hibiscus cannabinus L.). Tropical Plant Biol 15(4):261–275
Lin M et al (2023) Genome-Wide Identification and Transcriptional Analysis of the MYB Gene Family in Pearl Millet (Pennisetum glaucum). Int J Mol Sci 24(3):2484–2484
Liu Z et al (2023) Characterization of the REVEILLE family in Rosaceae and role of PbLHY in flowering time regulation. BMC Genom 24(1):49
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mahalakshmi V, Bidinger FR, Raju DS (1987) Effect of timing of water deficit on pearl millet (Pennisetum americanum ). Field Crops Res 15:327–339
Mengiste T et al (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15(11):2551–2565
Merga D (2020) Pearl millet (Pennisetum glaucum L.) breeding for adaptation and performance under drought condition: review. J Environ Earth Sci 10(4):1–10
Mistry J et al (2020) Pfam: the protein families database in 2021. Nucleic Acids Res 49(D1):D412–D419
Muthamilarasan M et al (2017) Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS ONE 9(10):e109920–e109920
Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79(4):639–648. https://doi.org/10.1016/0092-8674(94)90549-5
Penghui Li et al (2022) Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis and shoot development and stress response in tea plants (Camellia sinensis). Plant J 110(4):1144–1165
Perkins ML, Schuetz M, Unda F et al (2020) Dwarfism of high-onolignol Arabidopsis plants is rescued by ectopic LACCASE overexpression. Plant Direct. https://doi.org/10.1002/pld3.265
Rogozin IB et al (2012) Origin and evolution of spliceosomal introns. Biol Direct 7(1):11
Rosa T, Ayala Francisco J, Francisco R-T (2008) Alternative splicing: a missing piece in the puzzle of intron gain. Proc Natl Acad Sci USA 105(20):7223–7228
Sehgal D, Rajaram V, Armstead IP, Vadez V, Yadav YP, Hash CT, Yadav RS (2012) Integration of gene-based markers in a pearl millet genetic map for identification of candidate genes underlying drought tolerance quantitative trait loci. BMC Plant Biol 12:9
Stracke R, Werber M, Weisshaar B (2001) The R2R3–MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Su P, Hu J, Li Y et al (2018) The R2R3 MYB transcription factor mdmyb30 modulates plant resistance against pathogens and salt stress in apple. Plant Cell Environ 41(4):485–498. https://doi.org/10.1111/pce.13114
Sun M, Huang D, Zhang A, Khan I, Yan H, Wang X, Zhang X, Zhang J, Huang L (2020) Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol 20:323
Tombuloglu H, Kekec G, Sakcali MS, Unver T (2013) Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol Gen Genomics 288(3–4):141–155. https://doi.org/10.1007/s00438-013-0740-1
van Oosterom EJ, Mahalakshmi V, Bidinger FR, Rao KP (1996) Effect of water availability and temperature on the genotype- by-environment interaction of pearl millet in semi-arid tropical environments. Euphytica 89:175–183
Varshney Rajeev K et al (2017) Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 35(10):969–976
Wang D et al (2010) KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinform 8(1):77–80
Wang M, Liang C, Li C et al (2018) A wheat R2R3-MYB Gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. Front Plant Sci 9:422. https://doi.org/10.3389/fpls.2018.00422
Wang C, Wang H, Li P et al (2020) Developmental programs interact with ABA to coordinate root suberization in Arabidopsis. Plant J. https://doi.org/10.1111/tpj.14920
Wang Y et al (2021) Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol 21(1):328
Wei S, Hu W, Deng X et al (2016) Overexpression of soybean 1R-MYB1 confers tolerance to salt stress and improves growth in Arabidopsis thaliana. Front Plant Sci 7:1464. https://doi.org/10.3389/fpls.2016.01464
Xiangbo Z et al (2007) Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis:Circadian-related transcription factor CIR1. Plant J 51(3):512–525
Xiong A-S et al (2008) Gene duplication and transfer events in plant mitochondria genome. Biochem Biophys Res Commun 376(1):1–4
Yan H, Sun M, Zhang Z et al (2023) Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nat Genet. https://doi.org/10.1038/s41588-023-01302-4
Yang B et al (2018) RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet 14(12):e1007839
Yates AD et al (2021) Ensembl Genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res 50(D1):D996–D1003
Yuhui S et al (2020) E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. New Phytol 227(2):455–472
Yupeng W et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acid Res 40(7):e49
Zhang Z et al (2012) ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun 419(4):779–781
Zhang S et al (2022) Chromosome-scale genome assembly provides insights into speciation of Allotetraploid and massive biomass accumulation of elephant grass (Pennisetum purpureum Schum). Mol Ecol Res 22(6):2363–2378
Zhang H et al (2022) Genome-wide characterization, identification and expression profile of MYB transcription factor gene family during abiotic and biotic stresses in mango (Mangifera indica). Plants 11(22):3141–3141
Zhao Bo et al (2022) MYB44-ENAP1/2 restricts HDT4 to regulate drought tolerance in Arabidopsis. PLoS Genet 18(11):e1010473
Zihang He et al (2021) UNFERTILIZED EMBRYO SAC 12 phosphorylation plays a crucial role in conferring salt tolerance. Plant Physiol 188(2):1385–1401
Acknowledgements
This work was supported by the Modern Agricultural Industry System Sichuan Forage Innovation Team (no. SCCXTD-20201-16 to L.H.), the Sichuan Province Research Grant (no.2021YFYZ0013 to L.H.) and the National Natural Science Foundation of China(nos. 31771866 and 32071867 to L.H.).
Author information
Authors and Affiliations
Contributions
JL and YX were responsible for the experimental design and involved in all work, XD was responsible for the experimental data analysis and alignment, XM, JZ, CM, MS, YJ and LH suggested and revised the content of the article. All authors reviewed the manuscript and contributed to the article.
Corresponding author
Ethics declarations
Competing interests
This manuscript describes original work and is not under consideration by any other journal. All authors have approved the manuscript for submission and declare no potential competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lv, J., Xu, Y., Dan, X. et al. Genomic survey of MYB gene family in six pearl millet (Pennisetum glaucum) varieties and their response to abiotic stresses. Genetica 151, 251–265 (2023). https://doi.org/10.1007/s10709-023-00188-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-023-00188-8