Abstract
Connexins (Cxs) were first identified as subunit proteins of the intercellular membrane channels that cluster in the cell communication structures known as gap junctions. Mutations in the gap junction β2 (GJB2) gene encoding connexin 26 (Cx26) have been linked to sporadic and hereditary hearing loss. In some cases, the mechanisms through which these mutations lead to hearing loss have been partly elucidated using cell culture systems and animal models. The goal of this study was to re-assess the pathogenic roles of the GJB2 mutations by combining comparative evolutionary studies. We used Bayesian phylogenetic analyses to determine the relationships among 35 orthologs and to calculate the ancestral sequences of these orthologs. By aligning sequences from the 35 orthologs and their ancestors and categorizing amino acid sites by degree of conservation, we used comparative evolutionary methods to determine potential functionally important amino acid sites in Cx26 and to identify missense changes that are likely to affect function. We identified six conserved regions in Cx26, five of which are located in the Connexin_CCC, and another is in the connexin super family domain. Finally, we identified 51 missense changes that are likely to disrupt function, and the probability of these changes occurring at hydrophilic amino acid residues was twice that of occurring at hydrophobic residues in the trans-membrane regions of Cx26. Our findings, which were obtained by combining comparative evolutionary methods to predict Cx26 mutant function, are consistent with the pathogenic characteristics of Cx26 mutants. This study provides a new pathway for studying the role of aberrant Cx26 in hereditary hearing loss.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hereditary hearing loss is a very heterogeneous sensory deficit with different patterns of inheritance and a multitude of different genes (Smith et al. 2005) involved. Approximately 80 % of all cases of hereditary nonsyndromic hearing loss (NSHL) show autosomal-recessive inheritance; additionally, 15–20 % are autosomal-dominant, and approximately 1 % are linked to X-chromosome or mitochondrial DNA mutations. In the nuclear genome, approximately 140 deafness loci were mapped, and 66 genes for monogenic NSHL were identified (Van Camp and Smith 2013). The most frequent cause of nonsyndromic autosomal recessive hearing loss in humans is the mutations in the GJB2 (gap junction β2) gene encoding connexin 26 (Cx26), which is the transmembrane protein involved in the formation of connexins (Cxs). In the human inner ear, Cx26 has also been found to be highly expressed, and its crucial role in organ physiology has been revealed by its implication in different forms of hereditary hearing loss. Therefore, the mutations in human Cx26 have been closely linked to hereditary deafness.
Over 200 deafness-causing mutations and several polymorphisms and sequencing variants whose role in the pathogenesis of hearing loss is still unclear have been reported (Martínez et al. 2009) in the GJB2 gene to date. The spectrum and frequencies of GJB2 gene mutations have been characterized by significant interpopulation differences (Estivill et al. 1998; Azaiezr et al. 2004); however, due to the diversity of mutations and because novel mutations are continuously found in the GJB2 gene, the pathogenic role of different mutations of the gene and the structural properties of the protein remain largely unknown, making it difficult to predict the consequences of these mutations. Based on the diversity of mutations in the GJB2 gene and the continuous discoveries of new mutations, predicting pathogenic mutations and their correlation to disease phenotypes has become an important scientific endeavor. In this study, we explored GJB2 molecular structure characteristics and determined pathogenic missense mutations from known missense mutations in a molecular evolutionary direction.
Materials and methods
Data sources and phylogenetic analyses
Cx26 amino acid and nucleotide sequences for 35 species were extracted from Ensembl (http://asia.ensembl.org/index.html) (Table S1). The amino acid sequences were aligned using CLUSTALW 2.0 (Thompson et al. 1994, 2002).
MRBAYES 3.2.1 (Ronquist et al. 2012; Huelsenbeck et al. 2001) was used to construct a phylogenetic tree of GJB2 evolution by using amino acid of 35 species and Lamprey that is distantly related to human as an outgroup (Fig. 1). The Bayesian approach was used to combine the prior probability of a phylogeny with the likelihood of producing a posterior probability distribution in trees, and the posterior probability can be interpreted as the probability that the tree is correct (Huelsenbeck et al. 2001). We used MCMC algorithm to calculate posterior probabilities for each branch (Arvestad et al. 2003).
A Bayesian phylogenetic tree of GJB2. The numbers adjacent to the internal nodes represent the posterior probability that a clade is correct based on a consensus of 8,000 trees with approximately equivalent likelihood. Species’ categories are identified by different colours: red, primates; blue, carnivore; purple, artiodactyla; yellow, rodentia; green, marsupialia; orange, lagomorpha. The ancestral sequences of these species and mammal were calculated. The nattier blue represents non-mammals
As the evolutionary model, we used the GTR model with gamma-distributed rate variation across sites and a proportion of invariable sites, and we set the prior for the amino acid model to “mixed” to explore all of the fixed-rate models in MrBayes and achieve the most appropriate model. The analysis was started from random trees for four simultaneous and independent chains, including three hot chains and one cold chain. The analysis was run for 1,000,000 generations to ensure that we could achieve the lowest and stable average standard deviation, which is the convergency criteria of our analysis. Every hundredth tree was saved; the first 20 % of saved trees were classified as “burn-in” and were discarded. After ≈500,000 generations, stable likelihood estimates were achieved.
Conserved regions analyses
Homologous amino acid sites were divided into three categories: fixed, conservative and non-conservative sites. We used one-sample run tests (two-tailed) to determine whether fixed or conservative residues were associated. We defined as “conserved regions” those portions of the alignment that began and ended with fixed sites and comprised more than 80 % of such sites. Conserved regions of the gene were identified by using a sliding window of 5 aa. We compared levels of amino acid conservation both among the species themselves and among the sequences derived for their ancestors, and these ancestral sequences were calculated using a Bayesian phylogenetic analysis in which clades of sister taxa were constrained on the 1,000,000-generation consensus tree (Huelsenbeck and Ronquist 2001; Huelsenbeck and Bollback 2001).
Missense changes analyses
We analyzed the relationship between the distribution of conserved sites in the 35 species and their ancestors and that of some missense changes reported in the CRG (Center for Genomic Regulation) database (http://davinci.crg.es/deafness/index.php) and HGMD (The Human Gene Mutation Database, http://www.hgmd.org/) to determine the pathogenic missense changes of the GJB2 gene that are most likely to affect function in humans. Studying non-conservative substitutions at fixed or conservative sites and conservative substitutions at fixed sites can often yield significant insights. We used the Gonnet matrix (“G”) to identify missense changes involving non-conservative substitutions and the extent to which sites in the GJB2 sequence were fixed between 35 mammal sequences and ancestral sequences (“A”) (A&G method). We compared these predictions with those derived from the program SIFT, which estimates the degree of conservation by calculating the probabilities for all possible amino acids at each position in the alignment based on the sequences that are homologous to the query sequence, predicts a substitution to affect protein function, and generates the so-called SIFT score. We used the Chi-square test to evaluate the associations of conservative or non-conservative missense changes with fixed or conservative amino acid sites and with conserved regions.
We used SOSUI (Hirokawa et al. 1998), a classification and secondary structure prediction system for membrane proteins, to analyze the trans-membrane structure and amino acid properties for the Cx26 protein. The associations of missense changes with amino acid properties were also tested using the Chi-square test.
Results
Phylogeny of GJB2
There is a significant amount of variation in the sequence length of GJB2 among mammals, which resulted in an alignment of 226 codons for 31 eutherian mammals, the Anole lizard, the Lamprey, the Turkey and the Xenopus. Insertions which are phylogenetically uninformative, such as codons 222–246 of Lamprey, codons 227–248 of Xenopus and codons 230–263 of Turkey and so on, were removed from the phylogenetic analysis. In the phylogenetic tree of GJB2 (Fig. 1), all but 4 of the 19 (21 %) clades resolved with posterior probabilities >0.60.
A total of 165 (73.0 %) of the 226 human amino acid residues are fixed among mammals, and another 43 (19.0 %) are conservative. In contrast, 209 residues (92.5 %) are fixed among humans and various ancestors (including the ancestors of artiodactyla, primates, carnivora, rodentia, mammals, marsupialia and lagomorpha). During human evolution, ten conservative substitutions and a non-conservative substitution which affected the codon 162 that replaced Phe by Ser occurred in the mammals’ ancestor. A marked difference exists among the non-mammals and human GJB2, namely, only 119 (52.7 %) of the 226 human amino acid residues are fixed. We define these sites as “highly fixed” sites (HF sites) in which residues remain fixed among 35 species and their ancestors (Fig. 2b). Thus, 111 HF sites were identified, and they are not randomly distributed across human Cx26 amino acid sequence (z = −2.391, P < 0.02). A total of 34.5 % of the HF sites were identified across codons 146–213 (χ2 = 5.989 df = 1, P < 0.02), an interval that includes a gap junction channel protein cysteine-rich domain (Connexin_CCC), and 57.3 % were located in codons 2–108 that is the so-called connexin super family domain (Kar et al. 2012). The two domains which we can retrieve from the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/cdd/) are recognized conserved domains in the Cxs family. Besides, these HF sites also include 40 of 49 sites which are conserved within the connexin family and mutations of these residues are associated with deafness and skin disease (Figure S2) (Maeda et al. 2009). Six conserved regions in which amino acid identities are also above 80 % and whose lengths are 5–24 residues were also identified. Five of them are located in the Connexin_CCC and another is in the connexin super family domain (Figure S2).
a Alignment of Cx26 amino acids for human and 31 mammals. Markings under the alignment indicate the level of sequence conservation in all 15 eutherian mammals: asterisks, fixed residues; colon and dot, sites that include conservative substitutions from only strongly conserved or weakly conserved amino acid groups, respectively, based on the Gonnet matrix. asterisks represents the fixed residues, colon and dot represent the conservative residues, and others are non-conservative residues. b Alignment of Cx26 amino acids for 35 species and ancestral sequences derived from the phylogenetic tree in Fig. 1. The 111 HF sites in which residues remain fixed in this alignment are marked by orange
Pairwise comparisons between humans and other eutherian mammals reveal high levels of amino acid identity (Table S1). The average pairwise conservation is 98.3 ± 0.6 % between humans and other primates, and this value remains high between humans and rodents (92.9 ± 0.9 % on average). However, conservation is lower between humans and Lamprey or Xenopus (62.0 and 73.0 %, respectively).
Missense changes
From the CRG database and the HGMD, we retrieved 62 reported missense changes that are relevant to hereditary NSHL and that occur at 50 sites in GJB2. These changes are randomly distributed across the human Cx26 amino acid sequence (z = 0.430, P > 0.65) and across fixed or conservative sites in the 31 eutherian mammals studied (χ2 with correction for continuity: χ2 = 0.770, df = 1, P > 0.35).
Using the A&G method, we identified 51 of the 62 missense changes as likely to affect protein function (Table 1; Figs. 2a, b, 3). Forty-three of these changes, including 30 of the 39 missense changes predicted by SIFT, affect residues located in the HF sites. When the sequences of 31 eutherian mammals were compared, eight additional non-conservative changes occurred at fixed or conservative sites, including five of those 39 predicted by SIFT. Additional four changes (V95M, H100Y, H100L, L214P) predicted by SIFT are conservative changes that occur at conservative or fixed residues in the 31 eutherian mammals.
The three-dimensional structure of human Cx26. The orange dots respresent the positions of the mutations that we identified as likely to affect protein function. And the Cys residues that are essential for connexin stabilization through intramonomer disulfide bond formation (Cys53–Cys180, Cys60–Cys174 and Cys64–Cys169) are represented by pink sticks. Yellow region: N-terminal helix (NTH); lightblue regions: transmembrane domain (TM); green region: intracellular loop (CL); cyan regions: extracellular loops (E)
Using the 35 Cx26 amino acid sequences, we performed a similar comparison for 35 missense changes of known effect (Table S2). In this comparison, 26 of 30 changes known to be detrimental were correctly predicted to affect function; the remaining mutations (A40G, V95M, S113R and L214P) were conservative changes at conservative sites and was falsely classified as tolerated. Among five changes with no functional effect, V153I that is conservative changes at a non-HF site due to non-conservative in the non-mammals sequences were correctly predicted to be tolerated, but four (V27I, E114G, R127H and I203T) were incorrectly predicted to be detrimental (Table S2). Thus, the false-positive rate for Cx26 was 11.4 % (4/35), and the false-negative rate was 11.4 % (4/35). When using the SIFT prediction, the false-positive rate was 2.9 % (1/35), and the false-negative rate was 28.6 % (10/35).
The analysis result of SOSUI showed that the Cx26 protein comprised four trans-membrane regions and five non-transmembrane regions. Fifty-one of the changes predicted using the A&G method were identified at 41 sites in GJB2, and a statistical analysis did not contradict the conclusion that these sites were randomly distributed across transmembrane regions (22/41) and non-transmembrane regions (19/41) (χ2 = 2.856, df = 1, P > 0.05). In non-transmembrane regions, 19 of these 41 sites were randomly distributed across hydrophilic and hydrophobic amino acid residues (χ2 = 0.025, df = 1, P > 0.80). However, in transmembrane regions, 22 of these 41 sites were not randomly distributed (χ2 = 3.175, df = 1, P < 0.06). Among these sites, ten were hydrophilic and represented 38.4 % (10/26) of all hydrophilic amino acid residues in transmembrane regions; the remaining sites were hydrophobic and represented 18.2 % (12/66) (Fig. 4). We thus hypothesize that missense changes located in transmembrane regions are more likely to affect hydrophilic amino acid residues.
a The secondary structure of Cx26 protein, b the transmembrane helix figure for Cx26 protein. Boxes highlight residues where deleterious mutations predicted by the A&G method occur. Hydrophobic residues are black; neutral hydrophilic residues are blue; hydrophilic basic residues are blue bold; hydrophilic acidic residues are red bold
Discussion
In this study we reported the evolution characteristics of GJB2 in 35 orthologs, which showed a good consistency between the Bayesian tree and the Ensembl orthologous tree for GJB2 (Figure S1). Clades with posterior probabilities less than 0.6 are also poorly supported in the ML tree, including relationships among the orders primate, artiodactyla, carnivore, perissodactyla, Xenopus, microbat and shrew, which would be caused by the limited availability of Cx26 amino acid and nucleotide sequence resources for diverse species. The relationships among species in the tree imply that the molecular evolution of GJB2 essentially satisfies the basic rules of the species evolution.
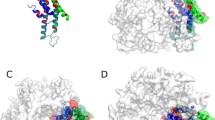
It is widely known that pathogenic mutations usually occur at main effect sites and regions in a gene sequence. These are highly conservative in the process of molecular evolution. Among six regions identified in Cx26, amino acid conservation across mammals and ancestral sequences was greater than 80 %. Region 6 is in the Connexin_CCC domain, and regions 1–5 are in the connexin super family domain. These two domains also exist in the other members of the Cxs family that can form transmembrane conduits for the exchange of small molecules and ions (Kar et al. 2012). Using Swiss-Model, a fully automated protein structure homology-modeling server, we successfully constructed the three-dimensional structures of 17 Cxs (including 6 beta-types, 9 alpha-types, 1 gamma-type and 1 delte-type) from the 20 known human connexin genes (Willecke et al. 2002). Through the three-dimensional structure comparisons between Cx26 and other Cxs, we obtained four conserved three-dimensional regions, which are codons 2–11, codons 15–98, codons 132–155 and codons 174–215 of Cx26 (Fig. 5a–c), and the six conserved regions are included in these four regions (Figure S2). Region 2, ranging from the codon 27–34, is located in the TM1 domain which is considered as the major pore-lining helix of Cx26; region 3–5 include the extracellular loop E1 and the N-terminal half of TM2, and region 6 includes the C-terminal half of E2 and the N-terminal half of TM4. The C-terminal half of E2 begins with a 310 turn and is followed by a conserved Pro-Cys-Pro motif that reverses its direction back to TM4, and E2 together with E1 forms the outside wall of the connexin (Fig. 3; Maeda et al. 2009). Region 1 located in the C-terminal half of NTH showed highly conservative in the phylogenetic analysis of Cx26, while showing very flexible in the multiple sequence alignment for human Cxs. We hypothesized that the conserved region 1 is peculiar to Cx26 and has some unknown function.
a The three-dimensional structure comparison between Cx26 and other beta-type Cxs; codons 2–98, codons 132–170 and codons 174–215 of Cx26 are conserved. Green: Cx26; marine: Cx25; magenta: Cx30.3; yellow: Cx30; pink: Cx31; wheat: Cx32. b The three-dimensional structure comparison between Cx26 and alpha-type Cxs; codons 2–11, codons 15–98, codons 132–155 and codons 162–215 of Cx26 are conserved. Green: Cx26; cyan: Cx31.9; magenta: Cx36; yellow: Cx37; orange: Cx45; wheat: Cx46; skyblue: Cx47; white: Cx50; slate: Cx59; pink: Cx62. c The three-dimensional structure comparison among Cx26, gamma-type Cx and delte-type Cx; codons 2–98, codons 132–156 and codons 162–215 of Cx26 are conserved. Green: Cx26; cyan: Cx31.3; pink: Cx40.1. Above all, the codons 2–11, codons 15–98, codons 132–155 and codons 174–215 of Cx26 are structural conserved regions among these 17 Cxs (considering that the intracellular loops and carboxyl terminus were just fantasy in the (Maeda et al. 2009) crystal structure and have not been resolved so far, and the C-terminus of Cx26 is the shortest in connexin family, we deleted them from these models in the structural comparisons)
The predicted pathogenic residues (C174R, R32C, R32L, R32H, Q80R, Q80P, E147R, S199F, A40E, W44S, W44C, W77R, R143W, R143Q, N206S, S139N, E47K, R75Q, R184W, W184P, R184Q, N54I and D179N) are mainly located in regions or residues critics for intra-protomer or interactions found by Maeda (Maeda et al. 2009), which can well explain why these mutations are predicted pathogenic by A&G, though some of these are predicted tolerated by SIFT.
The SIFT program identified 39 of 62 missense changes as affecting potentially functional residues in GJB2. We identified 35 of these 39 mutations and an additional 16 changes by using the A&G method. We obtained evidence of the pathogenic role for these 16 missense changes. The R165W mutation led to a constriction of the channel pore with no dye coupling in the intercellular dye-transfer experiment (Xiao et al. 2011). M163V has been reported to lead to failure of the homotypic junctional channel formation and the E101G change alters polarity of the cytoplasmic loop of Cx26, which would be expected to affect pH-dependent channel gating (Bruzzone et al. 2003; Jun et al. 2000). In in vitro functional studies, the M163L mutant Cx26 is defective in its ability to traffic to the plasma membrane and was associated with increased cell death (Stong et al. 2006; Matos et al. 2008). The S139N, N206S, E47K, V37I and L90V affected residues that are critical for the structure of Cx26 (del Castillo and del Castillo 2011). Additionally, an additional eight mutations (S19T, V37I, E47K, A88S, L90V, M93I, D179N and N206S) have been reported to be associated with the hereditary NSHL (Prasad et al. 2000; Wu et al. 2002; Joseph and Rasool 2009; Maeda et al. 2009), but the remaining three mutations (V27I, E114G and R127H) are wrongly predicted by the A&G method. The A&G method identified more changes because it considers the evolutionary relationship when identifying the fixed or conservative amino acid residues. SIFT compiles a dataset of functionally related protein sequences by searching a protein database using the PSI-BLAST algorithm and then builds an alignment of the homologous sequences with the query sequence. However, the low availability of species sequences for GJB2 led to the unsatisfactory search result that many members in the dataset are Cxs that are not related to hearing loss. In contrast, the A&G method recruited ancestral sequences of GJB2 to the dataset, which can improve the specificity of analysis. As a result, among the known detrimental missense changes in GJB2, the A&G method correctly predicted the functional effects of >85 %.
When analyzing trans-membrane regions of Cx26, we found that the probability of mutations occurring at hydrophilic amino acid residues was twice that of mutations occurring at hydrophobic residues (38.4 vs 18.2 %). A possible reason for this result is that mutations affecting hydrophilic residues more easily influence the stability of trans-membrane channel and transport function.
References
Arvestad L, Berglund AC, Lagergren J, Sennblad B (2003) Bayesian gene/species tree reconciliation and orthology analysis using MCMC. Bioinformatics 19(Suppl 1):i7–i15. doi:10.1093/bioinformatics/btg1000
Azaiezr H, Chamberlin GP, Fischer SM, Welp CL, Prasad SD, Taggart RT, del Castillo I, Van Camp G, Smith RJ (2004) GJB2: the spectrum of deafness-causing allele variants and their phenotype. Hum Mutat 24:305–311. doi:10.1002/humu.20084
Bruzzone R, Veronesi V, Gomès D, Bicego M, Duval N, Marlin S, Petit C, D’Andrea P, White TW (2003) Loss-of-function and residual channel activity of connexin26 mutations associated with non-syndromic deafness. FEBS Lett 533:79–88. doi:10.1016/S0014-5793(02)03755-9
del Castillo FJ, del Castillo I (2011) The DFNB1 subtype of autosomal recessive non-syndromic hearing impairment. Front Biosci 16:3252–3274. doi:10.2741/3910
Estivill X, Surrey S, Rabionet R, Melchionda S, D’Agruma L, Mansfield E, Rappaport E, Govea N, Milà M, Zelante L, Gasparini P (1998) Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet 351:394–398. doi:10.1016/S0140-6736(97)11124-2
Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379. doi:10.1093/bioinformatics/14.4.378
Huelsenbeck JP, Bollback JP (2001) Empirical and hierarchical Bayesian estimation of ancestral states. Syst Biol 50:351–366
Huelsenbeck JP, Ronquist F (2001) MRBAYES, Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi:10.1093/bioinformatics/17.8.754
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314. doi:10.1126/science.1065889
Joseph AY, Rasool TJ (2009) High frequency of connexin26 (GJB2) mutations associated with nonsyndromic hearing loss in the population of Kerala, India. Int J Pediatr Otorhinolaryngol 73:437–443. doi:10.1016/j.ijporl.2008.11.010
Jun AI, McGuirt WT, Hinojosa R, Green GE, Fischel-Ghodsian N, Smith RJ (2000) Temporal bone histopathology in connexin 26-related hearing loss. Laryngoscope 110:269–275
Kar R, Batra N, Riquelme MA, Jiang JX (2012) Biological role of connexin intercellular channels and hemichannels. Arch Biochem Biophys 524:2–15. doi:10.1016/j.abb.2012.03.008
Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T (2009) Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458:597–602. doi:10.1038/nature07869
Martínez AD, Acuña R, Figueroa V, Maripillan J, Nicholson B (2009) Gap-junction channels dysfunction in deafness and hearing loss. Antioxid Redox Signal 11:309–322. doi:10.1089/ars.2008.2138
Matos TD, Caria H, Simões-Teixeira H, Aasen T, Dias O, Andrea M, Kelsell DP, Fialho G (2008) A novel M163L mutation in connexin 26 causing cell death and associated with autosomal dominant hearing loss. Hear Res 240:87–92. doi:10.1016/j.heares.2008.03.004
Prasad S, Cucci RA, Green GE, Smith RJ (2000) Genetic testing for hereditary hearing loss: connexin 26 (GJB2) allele variants and two novel deafness-causing mutations (R32C and 645-648delTAGA). Hum Mutat 16:502–508
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2, efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029
Smith RJ, Bale JF Jr, White KR (2005) Sensorineural hearing loss in children. Lancet 365:879–890. doi:10.1016/S0140-6736(05)71047-1043
Stong BC, Chang Q, Ahmad S, Lin X (2006) A novel mechanism for connexin 26 mutation linked deafness: cell death caused by leaky gap junction hemichannels. Laryngoscope 116:2205–2210. doi:10.1097/01.mlg.0000241944.77192.d2
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2: Chapter 2: Unit 2.3. doi: 10.1002/0471250953.bi0203s00
Van Camp G, Smith R Hereditary hearing loss. http://hereditaryhearingloss.org. (last update: 3 June 2013)
Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Güldenagel M, Deutsch U, Söhl G (2002) Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem 383:725–737. doi:10.1515/BC.2002.076
Wu BL, Lindeman N, Lip V, Adams A, Amato RS, Cox G, Irons M, Kenna M, Korf B, Raisen J, Platt O (2002) Effectiveness of sequencing connexin 26 (GJB2) in cases of familial or sporadic childhood deafness referred for molecular diagnostic testing. Genet Med 4:279–288. doi:10.1097/00125817-200207000-00006
Xiao Z, Yang Z, Liu X, Xie D (2011) Impaired membrane targeting and aberrant cellular localization of human Cx26 mutants associated with inherited recessive hearing loss. Acta Otolaryngol 131:59–66. doi:10.3109/00016489.2010.506885
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31171217) and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institution and the College Students’ Practice and Innovation Training Projects of Jiangsu Province (2012JSSPITP1025) to X. Cao and the Grant from Jiangsu Health Administration of China (LJ201120) to G.-Q. Xing.
Author information
Authors and Affiliations
Corresponding author
Additional information
X.-H. Han and Y. Fan contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, XH., Fan, Y., Wei, QJ. et al. Understanding of the molecular evolution of deafness-associated pathogenic mutations of connexin 26. Genetica 142, 555–562 (2014). https://doi.org/10.1007/s10709-014-9803-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-014-9803-4