Abstract
An association analysis on the genetic variability for silica concentration in rice hulls was performed using a “Mini-Core” set of 174 accessions representative of the germplasm diversity found in the USDA world collection of rice. Hull silica concentration was determined in replicated trials conducted in two southern states in the USA and was analyzed for its association with 164 genome-wide DNA markers. Among the accessions, the average silica concentration ranged from 120 to 251 mg g−1. Ample variation was seen within each of the five sub-populations of rice, as well as the 14 geographic regions that the accessions originated from. There was also an effect due to location and accession × location (G × E) interaction demonstrating the importance of assessing silica concentration across multiple environments. Twelve markers on ten chromosomes were significantly associated with hull silica concentration. Six markers (RM5644, RM5371, RM1335, RM283, RM263, and RM178) corroborated quantitative trait locus for silica concentration identified in other mapping studies. Our results provide germplasm and genetic markers that will assist breeding efforts to develop cultivars that have either high or low hull silica concentration. High silica hulls are good raw material for silica based industrial compounds, while low silica hulls are more biodegradable.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is a silicon (Si) accumulating plant and, although Si is not an essential nutrient, it plays an important role in the growth and health of rice plants. Silicified cells are found in the epidermis of leaf blades and in the vascular tissues of the leaf sheath and stem (Ma and Takahashi 2002). Silica is found in varying concentrations throughout the plant and can make up to 10% of the shoot dry weight. Dai et al. (2005) reported that the silica concentration of two parental lines used in their mapping population had roughly twice the silica concentration in the hull than was present in the stem. Lombi et al. (2009) observed that the majority of the silica in the rice seed was deposited in the palea and lemma making up the hull.
Numerous studies have shown that Si in rice plant tissue increases along with an increase of Si application to soil (Datnoff et al. 2000; Kim et al. 2002; Rodrigues et al. 2004). In the 1930s and 1940s, Japanese researchers indicated that Si was effective in controlling some rice diseases (Takahashi 1995). Others have shown that an increase in plant Si significantly decreased the severity of diseases (Marschner 1995; Kim et al. 2002; Rodrigues et al. 2004). Silicon has been reported to increase yield up to 48%, reduce grain discoloration, control pests, and reduce the need for fertilizer (Correa-Victoria et al. 2001; Ishiguro 2001; Prabbu et al. 2001).
Genes responsible for silica uptake and transport have been identified in rice. Those genes regulate the total silica concentration in the rice plant as well as the distribution of silica concentration in specific tissues. Using conventional linkage mapping and mutant screening techniques, Ma and Yamaji (2006) identified the rice gene Low silicon rice1 (Lsi1). Lsi1 protein is a member of the aquaporin family and regulates silica concentration in the plant by functioning as an active silica influx transporter for silica uptake in root cells from the surrounding environment. Ma et al. (2007) later identified Lsi2, which regulates silica concentration in the plant by functioning as a silica efflux transporter that moves silica from the root cells into the stele. Using a homology search approach Yamaji et al. (2008) identified Lsi6, which also codes for a silica influx transporter and is a member of the aquaporin family. Lsi6 protein is active in the distribution of silica in the sheath and leaves of rice and functions by moving silica (silicic acid) out of the xylem into the surrounding xylem parenchyma cells.
While prior studies have examined silica concentration from a genetic perspective using conventional linkage mapping populations, mutant screening, and homology search techniques, there has been no association mapping study conducted to elucidate the genetic control of silica concentration in rice. Unlike linkage mapping studies, association mapping analyses, which take advantage of a whole genome scan approach, do not require prior knowledge of the genetic variation or the number of genes controlling the trait being analyzed. Association mapping can also be advantageous in its ability to examine hundreds, or even thousands, of varieties for marker-trait associations rather than being limited to the genetic constraints imposed when only two varieties are used in the creation of a mapping population (Zhu et al. 2008). This in turn can increase the probability of finding a novel gene or quantitative trait locus (QTL) that may not be present in the two parents of a conventional mapping population. Association mapping studies involving a wide range of markers and population sizes have been successful in identifying QTL for various traits (Sneller et al. 2009; Yu et al. 2009). In addition, association mapping can provide the fundamental knowledge necessary for developing conventional mapping populations desired for fine mapping specific QTLs or for direct use in breeding programs via marker assisted selection.
One of the issues confronting rice farmers and rice mills is how to dispose of the large amounts of abrasive, low nutritional value, high silica content rice straw and hulls. The US production of rough rice for 2010–2011 is estimated to be 11.4 billion kg (Childs and Baldwin 2010). Therefore, approximately 2.3 billion kg of hulls and 4.6 billion kg of straw will potentially be produced. Due to the high silica concentration, low density, high chemical stability, and low nutritional value of straw and hulls, the cost of transporting and disposal of these co-products is a major problem for the US rice industry. With the exception of California, most of the rice straw is burned in the field to facilitate its decomposition. However, burning rice straw is becoming less of an option due to environmental concerns. Some mills burn rice hulls to produce steam or electricity, which in turn makes disposal of the rice hull ash difficult due to the high silica concentration (70–95%) (Marshall 2004). Unused hulls and ash are usually taken to a landfill where they remain for years due to their chemical stability.
In an effort to reduce plant accumulation of Si, Ma et al. (2002) developed a rice mutant defective in Si uptake and found that the leaf blades were droopy, lodging increased, and photosynthesis decreased. Another approach for reducing the amount of hulls and ash going to the landfill is to use the silica for value-added products. Rice hulls have been used in such products as: particle board, poultry bedding, brick making, package cushioning, and absorbents. Due to the high silicon concentration, rice hulls and ash can also be used as raw materials in the production of silicon-based industrial materials with high economic value. These include silicon carbide, silica, silicon nitride, silicon tetrachloride, pure silicon, and zeolite (Sun and Gong 2001). As the silica in rice hulls is amorphous, it can be extracted at lower temperatures than silica derived from conventional manufacturing techniques, therefore, reducing the cost of production (Kalapathy et al. 2002).
Understanding the genetic control of silica concentration in rice will facilitate the development of new varieties with high silica concentration that will bring value to what is otherwise considered a waste product. For this to occur it is necessary to know the genetic variation of silica in rice hulls and to identify possible QTLs associated with silica concentration that can be used in breeding. The USDA world collection of rice contains over 18,000 accessions from 115 countries (Bockelman et al. 2003). In order to improve the efficiency of phenotypic and genotypic characterization, a representative Core subset (~1790 accessions) was developed using a stratified random sampling of the whole collection (Yan et al. 2007). From the Core subset, a “Mini-Core” collection (217 accessions) was selected using PowerCore software. Analysis of 26 phenotypic traits and 70 SSR markers proved that the Mini-Core represents the genetic diversity in the whole collection (Agrama et al. 2009). This Mini-Core collection includes representatives of all five sub-populations of rice; indica (IND), aus (AUS), temperate japonica (TEJ), tropical japonica (TRJ), and aromatic (ARO) as well as wild rice accessions (Li et al. 2010). The international origination of the Mini-Core accessions from 76 countries results in one of the most diversified collections in rice, which is valuable for mining genes of interest.
The objectives of this study were to (1) determine the growth stage and location of maximum silica deposition in the rice grain, (2) determine the genetic variability for silica concentration in the Mini-Core which is a representative subset of the USDA rice world collection; (3) determine the relationship of silica concentration with sub-population structure and geographic region of accession origin; and (4) determine the genetic markers associated with rice hull silica concentration using linkage disequilibrium mapping techniques.
Experimental
Materials (silica concentration in the rice grain)
Seeds of the cultivar “Wells” (PI 612439, http://www.ars-grin.gov/cgi-bin/npgs/acc/display.pl?1588913) and its recessive mutant for brittle culm, designated “Wells brittle” (GSOR 6, http://www.ars-grin.gov/cgi-bin/npgs/acc/display.pl?1651509), were planted in the greenhouse at the Dale Bumpers National Rice Research Center, Stuttgart, Arkansas (AR). Nomenclature (V1–V13, R0–R9) developed by Counce et al. (2000) was used to designate growth stages for plant sampling. At vegetative growth stage V2, twenty plants were transplanted into individual vented 2-liter pots filled with finely ground silt loam soil and moistened with water. The soil was kept moist and the plants were fertilized once a week with a commercial soluble fertilizer solution (15% N, 30% P2O5, 15% K2O, 0.02% B, 0.07% Cu, 0.15% chelated Fe, 0.05% chelated Mn, 0.0005% Mo and 0.06% water soluble Zn). Pots were arranged in a randomized complete block, split plot design experiment. Main plot treatments were the reproductive stages of development (R3, R4, R5, R6, R7 and R8) and subplot treatments were cultivar (Wells and Wells brittle). Pots within a subplot replication were rotated twice weekly to minimize position effect within the greenhouse. Beginning at vegetative growth stage V11 each plant was monitored daily for reproductive growth stage development. When a sufficient number of seeds of a designated growth stage were present on the plant, the panicles were harvested and the individual grains were removed and grouped according to reproductive growth stages R3 through R8. Silica concentrations were determined on the whole grains at each developmental stage. Because of the variation in development of grains along the length of the panicle and among tillers of the same plant, and in order to obtain enough sample for each reproductive stage, grains were harvested from plants in different pots and at different times within a subplot replication and bulked together. Since the weights of the seeds are different at each developmental stage (Fig. 1), the number of seeds needed for 0.2 g varied. Therefore, the data was reported on per grain basis, instead of per gram basis, derived by dividing the concentration by the number of seeds needed to make 0.2 g.
Wells and Wells brittle were also grown in field plots at University of Arkansas Rice Research and Extension Center near Stuttgart, AR in 2002. Rice seeds were sown in rows 19 cm apart, 500 seeds m2. The soil was a DeWitt silt Loam (Typic Albaqualfs). Rice was flooded at the V4 through V5 stages of development. Standard fertilization and weed control practices were used. Rice was drained for harvest 14 days after 50% heading. The rice plants were harvested by hand at R8, threshed, and aspirated. After drying overnight, rice grain for these analyses were allowed to equilibrate to approximately 12% moisture. Two aliquots of R8 seeds (0.2 g) from each replication (4) of each cultivar were analyzed for their silica concentration with and without the hulls and after milling to 10% (removal of bran layer) degree of milling.
Materials (Mini-Core)
One hundred and seventy-four accessions selected from the Mini-Core that originated from 14 global regions (Agrama et al. 2009, 2010) and representing six genetic groups (IND n = 54, AUS n = 37, TEJ n = 32, TRJ n = 26, ARO n = 5, and Admix n = 20) were grown in field plots near the Dale Bumpers National Rice Research Center, Stuttgart, AR and the Rice Research Unit (RRU), Beaumont, Texas (TX) in 2009. The accessions were selected based on those that would mature and set seed at both locations. Seed originating from a single plant selection of each accession was used for sowing. A randomized complete block design was used with two replications per location. The samples from each plot were harvested by hand, threshed and cleaned using standard field equipment (ALMACO, Nevada, Iowa), and dried to approximately 12% moisture using a forced air drier. Rough rice samples from the plots were dehulled with a Satake Rice Machine (Satake Engineering Co., LTD., Ueno Taito-Ku, Tokyo) and after drying at 80°C for 2 h, the hulls (~3 g) were stored in 50 ml polypropylene tubes (Cat. # 05-539-5, Fisher Scientific, Houston, TX) at room temperature (22°C) until analyzed.
Silica concentration
Silica was determined using the molybdenum yellow method described by Saito et al. (2005). Dried hulls (section “Materials (Mini-Core)”) or grains (section “Materials (silica concentration in the rice grain)”) (0.2 g) were placed in 15 ml polypropylene tubes and 10 ml of hydrofluoric acid (HF) solution (1.5 M hydrofluoric acid–0.6 M hydrochloric acid) was added. The samples were allowed to sit at room temperature (22°C) for 1 h swirling every 15 min. The solution from the hulls was diluted 1:1 with deionized water, and 100 μl of the diluted solution was place into a 15 ml centrifuge tube. To neutralize the acid and obtain the desired pH, 6 ml of 0.5 M boric acid and 2 ml of working molybdenum solution was added to each tube. After vortexing and allowing to sit at room temperature for 3 min., 1 ml of 0.5 M citric acid and 900 μl of water was added to each sample (total = 10 ml), vortexed and allowed to sit at room temperature for 10 min. The developed color was read at 400 nm using a Cary 50 spectrophotometer (Varian Australia Pty. Ltd., Mulgrave, Victoria, Australia) and converted to silica concentration according to Saito et al. (2005). Silica concentrations were expressed on a dry weight basis. All tubes were made of polypropylene and reagent blanks ran on each batch to ensure they contained no detectable silica. A reagent blank and standards were run with each set and each sample was run in duplicate.
X-ray diffraction
The structural analysis of the silica in the hull before, during, and after extraction was determined using x-ray diffraction (XRD) by adapting the method of Proctor (1990). A long grain rice cultivar, “Rondo” (Yan and McClung 2010), was planted in a field plot at the Dale Bumpers National Rice Research Center, Stuttgart, AR in 2009 and bulk harvested at maturity. After drying the rough rice to 12% moisture, grains were dehulled and the hulls dried as described above. The dried hulls (0.2 g) were extracted for 0, 15, 30, 45, and 60 min with HF solution (see above). After each extraction time the hulls were rinsed with water and dried at 130°C for 2 h. The processed hulls were characterized by x-ray diffraction using a Philips PW1830 model powder diffractometer (Philips, Almelo, The Netherlands) with Cu–Kα radiation (1.5419Å), a diffracted beam monochromator, ¼° divergence and 0.45 mm receiving slits. The silica concentration was determined semi-quantitatively by measuring the area under the associated peak at 22°, less the smooth continuous background determined using the PANalytical X’Pert HighScore software package (v. 2.2.5, PANalytical B.V., Almelo, The Netherlands).
Genotyping
One hundred and fifty-six SSR markers (marker sequence and genomic location information available from Gramene, http://www.gramene.org), two indel markers (RID 12, Sweeney et al. 2006; and Pi-ta, Wang et al. 2010), and six SNP markers (Waxy EX1 G/T, EX6, and EX10, Chen et al. 2010; and 3 ALK genes, SNPs, Bao et al. 2006) distributed across the rice genome at approximately every 10 cM were used to genotype 174 accessions from the Mini-Core collection. Markers sequences for AL606682-1, AP5652-1, AP5652-2, con673, and LJSSR1 are referenced in Li et al. 2011. The physical location (Mb) of the markers in the rice genome are shown in Fig. 4.
DNA from the leaf tissue of each Mini-Core accession was extracted using two methods, a CTAB method as described by Hulbert and Bennetzen (1991) and a rapid DNA extraction method described by Xin et al. (2003). PCR marker amplifications were performed in 25 μl volumes containing 20 ng of template DNA, and final concentrations of: 10 mM Tris–HCl at pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 300 nM each primer, and 1 unit of Taq DNA polymerase (Promega, Madison, WI, USA). For PCR with template DNA prepared using the rapid extraction method, bovine serum albumin and PVP40 were added at 0.1 and 1% concentrations relative to the final reaction volume, respectively. For each marker, the primer was labeled with a fluorescent dye (6FAM, NED or Hex; Applied Biosystems, Foster City, CA, USA or Integrated DNA Technologies, Coralville, IA, USA).
PCRs were performed in MJ Research thermal cyclers (Bio-Rad, Hercules, CA, USA). PCR amplification and analysis of SSR’s markers were performed as described in Costanzo et al. 2011.
Association mapping and genetic analysis
Ancestry relationship and sub-population structure among the Mini-Core accessions have been previously described (Agrama et al. 2010; Li et al. 2010) and, in part, verified again in this study. The software Structure 2.3.3 (Pritchard et al. 2000; Falush et al. 2003) was run with a burn in of 50,000 and run length of 80,000 to confirm population structure. Eight independent replications using sub-population numbers ranging from one to eight were run with the options of admixture (Admix) being present and correlated allele frequencies selected. Structure results were interpreted using the Evanno et al. (2005) method that examines the ad hoc statistic ΔK, which measures the rate of change in the log-likelihood value and provides an estimate of the population number. Principal components analysis (PCA) was performed with Nei’s genetic distance (Nei 1973) using GenAlEx 6.3 (Peakall and Smouse 2006) with 164 markers as an alternative to the program Structure to determine the population sub-structure (Q) of the lines. PowerMarker 3.25 (Liu and Muse 2005) was used to determine polymorphism information concentration (PIC values) of the markers and allele frequency. Rare genotypes occurring less than two percent of the time were removed from the data set prior to association mapping analysis. Frequency based distance was generated using the Rogers 1972 distance option and the subsequent results were used to create a UPGMA tree in PowerMarker. Mega 4 (Tamura et al. 2007) was used to graphically display the UPGMA tree. Kinship values were determined using SPAGeDI 1.3 (Hardy and Vekemans 2002) with the Loiselle et al. (1995) option.
Tassel 2.1 (http://www.maizegenetics.net/) was used to run association mapping. Two mapping models were compared; the mixed linear model (MLM) as described in Yu et al. (2006), which accounts for both population (Q) and kinship (K), and the general linear model, which accounts for Q only. Silica concentration was averaged across the two field replications at each location. The means of each location were analyzed both independently (AR and TX locations) and then averaged together to examine the overall effect (combined locations).
Statistical analysis of silica concentration
The silica concentration was analyzed with the GLIMMIX procedure in SAS (Version 9.2, SAS Institute, Cary, NC). The statistical model followed the experimental design of randomized complete block, with all effects considered as random (accessions, locations, and blocks). Within the GLIMMIX procedure, best linear unbiased predictors (BLUPs) and predicted standard errors were calculated using the SOLUTIONS option. Broad sense heritability estimates were calculated using the method of Singh et al. (1993).
Results and discussion
Silica location in rice kernels
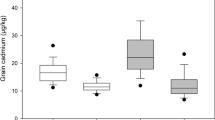
Figure 1 shows the grain weight, moisture concentration, and silica concentration of Wells Brittle and Wells at each developmental stage, as defined by Counce et al. (2000). Figure 1a, b show the individual grain weight (mg) and moisture concentration (%) of Wells brittle and Wells, respectively at each developmental stage. Wells brittle and Wells both gained weight at the same rate and the moisture profiles for each were also the same. The grain weight at R3 (panicle exertion) was 2 mg grain−1 and the weight increased slightly till R4 (anthesis). From R4 to R8, the grain weight dramatically increased to 23 mg grain−1 due to grain filling. The grain moisture concentration decreased during anthesis from 52% at R3 to 47% at R4 and then increased to 59% at R5 (caryopsis elongation) (Fig. 1b). Then, the moisture started steadily decreasing in the grain from R5 to R8 (grain dry-down) (Fig. 1b). During the R5 growth stage and early R6 (grain filling), the caryopsis is expanding and filling with starch, however, the contents are liquid or soft dough, and thus, have high moisture. The starch hardens during mid- and late-R6 as the moisture content begins to decrease and the grain continues to mature.
Figure 1c shows the silica concentration (mg grain−1) in the seeds of Wells brittle and Wells at each developmental stage. Unlike the moisture and grain weight, Wells brittle and Wells did not have the same silica profile. Both lines had <0.1 mg grain−1 silica at the R3 and up to ~0.2 mg grain−1 at the R5. There was very little increase in silica for Wells after R5. However, the silica concentration for Wells brittle went from 0.2 mg grain−1 at R6 to 0.4 mg grain−1 at R8, while it only increased to 0.25 mg grain−1 for Wells. Therefore, the accumulation of silica appears to be different for some genotypes after the R6 growth stage.
Figure 2 shows the silica concentration (mg grain−1) of rough, brown (the bran still on but without the hull), and milled (without the bran and hull) rice grains of Wells and Wells brittle at the R8 developmental stage. The majority of silica (75%) appeared to be in the hull. For Wells brittle and Wells the rough rice contained 0.40 and 0.33 mg grain−1, brown rice contained 0.08 and 0.01 mg grain−1, and milled rice contained 0.02 and 0.01 mg grain−1, respectively. Therefore, analysis of rice hulls at the R7 (physiological maturity) or later can be used to differentiate maximum silica accumulation in the grain among genotypes. The difference in silica concentration seen at R8 in the two sample sets (Figs. 1, 2) could be due to the environment in which they were grown. The first set was grown in the greenhouse and the second set was grown in the field.
X-ray diffraction analysis
XRD showed that, after digesting for 1 h, peaks were seen at 2θ = 15.0, 22.0 and 34.5° (Fig. 3). Although the peaks at 2θ = 15.0 and 34.5° remained approximately the same, the area under the peak at 2θ = 22.0° did not show significant reduction with extraction time up to 30 min. Indeed, the area under the peak at 2θ = 22.0° increased with extraction time in some cases. The 2θ = 22.0° peak is characteristic of disordered opal CT, α-cristobalite, which is indicative of amorphous silica (Proctor 1990). The area under the curve was found not to be representative of the concentration of extractable silica. Since the extraction process will remove components other than silica, the increase in the 2θ = 22.0° peak may be due to the difference in the relative concentration of the remaining silica to the undissolved material. Saito et al. (2005) found that the aforementioned method had a high correlation to the gravimetric method. They speculated that the slight increase in silica concentration seen in the gravimetric method may be due to incomplete dehydration or impurities. XRD shows that it may be due to a small amount of silica that is not extractable by HF. However, the amount of silica remaining is negligible and, therefore, the HF extraction can be used as an alternative to the gravimetric method.
X-ray Diffraction patterns of residual rice hulls obtained following digestion with HF for time periods of 0, 5, 15, 30 and 60 min, respectively. The intensities of the datasets are offset by arbitrary values to make viewing easier. The inset shows the integrated peak area above background for the peak at 2θ = 22°, which, for this series, increases with digestion time
Hull silica concentration analyses
Silica concentration of the Mini-Core averaged 200 mg g−1, and ranged from 120 mg g−1 ± 21.40 for ACNO 430909, an Admixture of aus (AUS), indica (IND) and aromatic (ARO) sub-population genotypes, to 251 mg g−1 ± 12.66 for ACNO 353722, an AUS sub-population accession, both from Southern Asia (Table 1). The non-Admix sub-population accession with the lowest silica concentration was ACNO 439683, a TEJ from Eastern Europe, having a silica concentration of 147 mg g−1 ± 4.47. Wide variation in silica concentration was observed in all genetic populations. The means of the TRJ accessions (219 mg g−1) and AUS accessions (208 mg g−1) were greater than the overall mean of the entire Mini-Core (200 mg g−1), whereas, other groups (TEJ, ARO, IND and Admix) had mean values less than the average of the Mini-Core. All the accessions native to Central America region (n = 9) except ACNO 2169, a TRJ, had higher silica concentration than the Mini-Core average, while the silica concentrations of accessions native to the Mideast (n = 5), Eastern Europe (n = 8), Central Asia (n = 9), and North America (n = 3) were below the Mini-Core average with a few exceptions. Some 25% of the Mini-Core was represented by accessions from Southern Asia (n = 44) which exhibited the widest range of silica concentration, 120–251 mg g−1 ± 5.17. The wide variation in silica concentration in rice hulls among the Mini-Core accessions, genetic populations, and geographic regions of the world demonstrates that there is considerable germplasm variability for breeders to utilize for modifying silica concentration in the development of new cultivars.
The variance due to genetics (i.e. accessions) accounted for 32.4% of the total variation for silica concentration, indicating that this trait has a moderately high level of heritability (48%). The silica concentration of samples grown in TX were lower than those grown in AR, with means of 187 mg g−1 ± 25.60 and 213 mg g−1 ± 26.12, respectively, with environment (i.e. location) accounting for 32.2% of the silica concentration variation. The cause of the location effect is most likely due to differences in soil composition or temperature during the growing season. However, the genetic × environment effect accounted for 25.6% of the total variation, highlighting the importance of considering GxE interactions when selecting accessions for breeding. Some accessions were identified as being stable across environments. Accessions ACNO 549253 and ACNO 373536, an AUS and IND from Southern Asia, respectively, were identified as having consistently high silica concentrations (228–236 and 220–237 mg g−1), respectively, whereas, accessions ACNO 154531, a TEJ from North Pacific, was consistently low (156–169 mg g−1) (Fig. 4).
Molecular sub-structure of Mini-Core collection
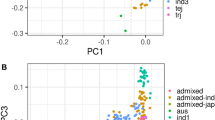
Structural analysis indicated that the Mini-Core collection could be classified into five sub-populations, consistent with the known structural studies in rice (Garris et al. 2005; Agrama et al. 2010). Only five ARO accessions were present in this study, and they formed a separate cluster in both the UPGMA tree and PCA. Twenty accessions were classified as admixtures, as was previously reported by Agrama et al. (2010), indicating that these accessions are a result of intermating between sub-populations. The PCA of marker allele variation explained 73.25% of the total genotypic variation in the Mini-core and confirmed the five main sub-populations identified by the Structure analysis. The UPGMA tree (Fig. 5) based on Roger’s genetic distance provided another way to visualize the genetic similarities of the accessions and showed similar results with both Structure and PCA. This information is useful to breeders for designing crosses to change silica concentration while maximizing/minimizing genetic variability in other genomic regions.
UPGMA tree of 174 lines genotyped with 164 molecular markers and clustered using Roger’s genetic distance. Five rice sub-populations are represented in individual clusters with the small group of aromatics (five lines) forming their own distinct cluster. Lines are labeled with their corresponding sub-population and ACNO number. Sub-populations are aromatic (ARO), aus (AUS), indica (IND), temperate japonica (TEJ), tropical japonica (TRJ), and admixtures (ADMIX)
Association mapping
Association mapping results of the mean silica concentration for the AR and TX samples were compared using both the GLM and MLM method of Tassel 2.1. The MLM method controls Type I and Type II error rates by accounting for both population structures (Q) and kinship (K) (Yu et al. 2006), while GLM only takes population structure into account. In this study the MLM method provided better control of the error rates, thus only results from the MLM are reported. Results of the MLM showed significant silica concentration associations (p ≤ 0.01) for four markers from AR of which three were detected in the combined analysis, four markers from TX of which none were detected in either the AR or combined analysis, and seven markers from the combined study including three markers seen in AR and four markers unique to the combined analysis (Table 2; Fig. 4). Marker effects were generated for the major alleles (alleles having the greatest effect on increasing or decreasing the silica concentration) of significant markers (Table 2). Rare alleles making up 2% or less of the number of observations were discarded from analysis. In this study, all of the markers in which ten or fewer of the major alleles had been observed in a homozygous condition were either validated by previous QTL mapping studies, or in the case of RM484, by nearby markers.
Markers identified in the association mapping study differed between the AR and TX locations (p ≤ 0.01). When coupled with the variance associated with the genetic × environment interaction (25.3%), this highlights the importance of environmental factors on silica uptake. The amount of silica present in the soil, the presence of other elements and/or nutrients, the amount of light, and temperature are all factors known to affect silica concentrations in the plant (Ma and Takahashi 2002; Ma et al. 2002). It is conceivable that different genes and/or gene pathways influenced by different environmental variables are involved in regulating the silica concentration of the plant. In order to account for possible environment effects, and identify where silica concentration is significantly associated with markers in both environments, data of silica concentrations from the AR and TX locations were combined and a third association mapping analysis was conducted.
In addition to the known silica transporter genes Lsi1 (chromosome 2 at 29.83 Mb), Lsi2 (chromosome 3 at 1.11 Mb), and Lsi6 (chromosome 6 at 6.65 Mb), other studies have reported numerous QTLs involved with silica concentration. The mapping study performed by Wu et al. (2006) found a QTL for silica concentration in rice sheaths and leaves spanning a large region on chromosome 7 from 16.9 to 27.1 Mb, which is near RM1335 (chromosome 7, 28.3 Mb), the marker with the highest significance in the TX study. Similar conventional mapping studies by Dai et al. (2005, 2008) identified QTLs for rice hull silica concentration within 1.5 Mb of the marker-trait associations identified in our association mapping studies. They reported a QTL on chromosome 1 between 5.6 and 5.8 Mb which is in close proximity to the marker RM283 (chromosome 1 at 4.9 Mb) from our combined study. Another silica concentration QTL on chromosome 5 from 25 to 25.6 Mb reported by Dai et al. (2005) positionally overlaps with RM178 (chromosome 5 at 25.1 Mb) from our TX study. The major allele of RM178 (114 bp) is present in over 40% of the lines tested and had a strong negative effect on silica concentration, lending further support for its involvement with silica concentration in the hull. RM 263 (chromosome 2 at 25.9 Mb) from the AR study, RM6544 (chromosome 11 at 3.9 Mb) from the AR and combined analyses, and RM5371 (chromosome 6 at 25.8 Mb) from the TX study all occur within a 1.5 Mb region that Dai et al. (2005) had previously identified as having additive by additive QTL effects.
Additionally, RM5371 which has a major allele with a negative effect (−18.6 mg g−1) on hull silica concentration, falls within a QTL region identified for arsenic concentration in rice grains (Zhang et al. 2007). Silica uptake is known to be involved with both arsenic and phosphorus uptake in rice (Guo et al. 2007). High arsenic concentration in rice grain is associated with health concerns in certain areas of the world. Understanding the relationship between arsenic and silica deposition in the grain could become an important step in overcoming high arsenic concentration in the grain. The marker trait-association for silica concentration in the hulls found in this study coupled with the arsenic concentration QTL found by Zhang et al. 2007, suggests that the region near RM5371 could be promising for further study.
Marker RM5953, which was not significant at the TX location, was the most significant marker (p value = 0.00086) in the combined analysis and was significant in the AR study. The major allele of RM5953 (111 bp) had a 14.5 and 15.3 mg g−1 marker effect in the combined and the AR study, respectively, showing a strong positive effect for increased silica concentration associated with this allele. RM23869 was the second most significant marker-trait association in the combined analysis and the most significant marker-trait association in the AR study (p = 0.00023). The major allele (182 bp) was well supported by 37 homozygous lines and had a negative effect on silica concentration in both locations. RM6544 was another marker that was detected in both the combined analysis and AR study, having a major allele (165 bp) with positive effect.
Four markers, RM283, RM489, RM171, and RM484 were unique to the combined analysis. RM283, a marker less than 1 Mb away from a previously identified silica QTL region on chromosome 1 (Dai et al. 2005), had a major allele with a large negative effect on silica concentration (−19.7 mg g−1), but was found homozygous in only 5 accessions. RM489 on chromosome 3 had a major allele with 25 homozygous observations and a positive effect on silica concentration of 15.2 mg g−1. Two other markers of interest in the combined study were RM171 (chromosome 10 at 19.1 Mb) and RM484 (chromosome 10 at 21.1 Mb). Both RM171 and RM484 flank the rice OsNIP3;1 gene (chromosome 10 at 19.7 Mb). OsNIP3;1, like the previously identified rice silica transport genes Lsi1 and Lsi6, is a member of the aquaporin protein superfamily. OsNIP3;1 is expressed in the roots, inflorescence and seeds of rice (Liu et al. 2009). Mitani et al. (2008) expressed OsNIP3;1 in Xenopus oocytes and found it to function as a transporter although it was incapable of transporting silicic acid in their system. Evidence that OsNip3;1 plays a role in silica accumulation in grain hulls has yet to be reported. The major alleles of RM171 and RM484 both had strong negative effects on silica concentration, −18.7 and −22.4 mg g−1, respectively (Table 2). While the major allele of RM484 had only four homozygous observations, RM171 had 42 homozygous observations as well as a strong negative effect. These two markers occur in close proximity to each other, within 2 Mb, lending support to a potential QTL being present in this region. The marker-trait associations provided by this association mapping study should provide a starting point to look for additional silica concentration controlling genes or QTLs as well as to develop rice cultivars with high or low hull silica concentration.
Conclusions
The silica concentrations present in rice hulls varied across all five sub-populations of rice. Eighty-five percent of TRJ accessions had higher silica concentrations than the Mini-Core mean, indicating that TRJ is a rich genepool for increasing rice silica concentration. Seventy-five percent of the USA rice production is for the long grain market, with essentially all of this germplasm derived from TRJ. Agronomically acceptable rice cultivars with high silica concentration would increase the value of the hulls as they could be utilized for the production of silica based industrial compounds. Silica concentration also varied among accessions from different geographic regions. Most of the accessions from Central America had higher silica concentrations while those from the Mideast and Eastern Europe had lower silica concentrations as compared to the Mini-Core mean. Accessions from these regions may offer germplasm that could be used to develop rice varieties that extract less silica from the soil. Decreased silica concentrations would aid in the biodegradation of waste hulls and straw however, negative effects on disease resistance or lodging susceptibility would have to be tested. Environment effects were an important factor in the association mapping analyses, highlighting the importance of GxE interactions and the necessity of having multiple test sites for silica evaluation. In our study four significant marker-trait associations were found at both the AR and the TX locations and seven marker-trait associations were found in the combined analysis of AR and TX. Five of the marker-trait associations found in this study are within 1.5 Mb of reported QTLs for silica concentrations from linkage mapping studies, and one marker-trait association (RM5371 on chromosome 6 at 25.83 Mb) overlaps with a QTL involved in grain arsenic concentration as well as silica concentration. Association mapping of the diverse germplasm in the USDA rice Mini-Core collection was an effective method for identifying new genetic markers and validating previously reported marker regions associated with silica concentration.
References
Agrama HA, Yan W, Lee F, Fjellstrom R, Chen M-H, Jia M, McClung AM (2009) Genetic assessment of a mini-core subset developed from the USDA rice genebank. Crop Sci 49:1336–1346
Agrama HA, Yan WG, Jia MH, Fjellstrom R, McClung A (2010) Genetic structure associated with diversity and geographic distribution in the USDA rice world collection. Nat Sci 2:247–291
Bao JS, Corke H, Sun M (2006) Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physiochemical properties in rice (Oryza sativa L.). Theor Appl Genet 113:1171–1183
Bockelman HE, Dilday RH, Yan WG, Wesenberg DM (2003) Germplasm collection, preservation and utilization. In: Smith CW, Dilday RH (eds) Rice origin, history, technology and production. Wiley, New York, pp 597–625
Chen M-H, Fjellstrom RG, Christensen EF, Bergman CJ (2010) Development of three allele-specific codominant rice Waxy gene PCR markers suitable for marker-assisted selection of amylose content and paste viscosity. Mol Breed 26:513–523
Childs N, Baldwin K (2010) Rice Outlook. July 12. USDA Economic Research Service. http://usda.mannlib.cornell.edu/usda/ers/RCS//2010s/2010/RCS-07-12-2010.pdf. Accessed 27 April 2011
Correa-Victoria FJ, Datnoff LE, Okada K, Friesen DK, Sanz JI, Snyder GH (2001) Effects of silicon fertilization on disease development and yields of rice in Colombia. In: Datnoff LE, Snyder GH, Korndorfer GH (eds) Studies in plant science 8: silicon in agriculture. Elsevier Science B.V, Amsterdam, pp 313–322
Costanzo S, Jackson AK, Brooks SA (2011) High-resolution mapping of Rsn1, a locus controlling sensitivity of rice to a necrosis-inducing phytotoxin from Rhizoctonia solani AG1-IA. Theor Appl Genet 123:33–41
Counce PA, Keisling TC, Mitchell AJ (2000) Uniform adaptive and objective system for expressing rice development. Crop Sci 40:436–493
Dai W-M, Zhang K-Q, Duan B-W, Zheng K-E, Zhuang J-Y, Cai R (2005) Genetic dissection of different organs in rice. Crop Sci 45:1345–1352
Dai W-M, Zhang K-Q, Wu J-R, Wang L, Duan B-W, Zheng K-L, Cai R, Zhuang J-Y (2008) Validating a segment on the short arm of chromosome 6 responsible for the genetic variation in the hull silicon content and yield traits of rice. Euphytica 160:317–324
Datnoff LE, Victoria-Correa FJ, Seebold KW, Snyder GH (2000) Silicon management of blast in upland and irrigated rice ecosystems. In: Tharreau D, Lebrun MH, Talbot NJ, Notteghem JL (eds) Advances in rice blast research. Kluwer Academic, Amsterdam, pp 180–187
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics 164:1567–1587
Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Guo W, Zhu YG, Liu WJ, Liang YC, Geng CN, Wang SG (2007) Is the effect of silicon on rice uptake of arsenate (AsV) related to internal silicon concentrations, iron plaque and phosphate nutrition? Environ Pollut 148:251–257
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hulbert SH, Bennetzen JL (1991) Recombination at the Rp1 locus of maize. Mol Gen Genet 226:377–382
Ishiguro K (2001) Review of research in Japan on the roles of silicon in conferring resistance against rice blast. In: Datnoff LE, Snyder GH, Korndorfer GH (eds) Studies in plant science 8: silicon in agriculture. Elsevier Science B.V, Amsterdam, pp 277–291
Kalapathy U, Proctor A, Shultz J (2002) An improved method for production of silica from rice hull ash. Biores Technol 85:285–289
Kim SG, Kim KE, Park EW, Choi D (2002) Silicon-induced cell wall fortification of rice leaves: a possible cellular mechanism of enhanced host resistance to blast. Phytopathology 92:1095–1103
Li X, Yan W, Agrama H, Hu B, Jia L, Jia M, Jackson A, Moldenhauer K, McClung A, Wu D (2010) Genotypic and phenotypic characterization of genetic differentiation and diversity in the USDA rice mini-core collection. Genetica 138:1221–1239
Li X, Yan W, Agrama H, Jia L, Shen X, Jackson A, Moldenhauer K, Yeater K, McClung A, Wu D (2011) Mapping QTLs for improving grain yields using the rice mini-core collection. Planta. doi:10.1007/s00425-011-1405-0
Liu K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Liu Q,Wang H, Zhang Z, Wu J, Feng Y, Zhu Z (2009) Divergence in function and expression of the NOD26-like intrinsic proteins in plants. BMC Genomics. doi:10.1186/1471-2164-10-313
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lombi E, Scheckel KG, Pallon J, Carey AM, Zhu YG, Meharg AA (2009) Speciation and distribution of arsenic and localization of nutrients in rice grains. New Phytol 184:193–201
Ma JF, Takahashi E (2002) Soil, fertilizer, and plant silicon research in Japan. Elsevier Science B V, Amsterdam
Ma JF, Yamaji N (2006) Silicon uptake and accumulation in higher plants. Trends Plant Sci 11:392–397
Ma JF, Tamai K, Ichii M, Wu GF (2002) A rice mutant defective in Si uptake. Plant Physiol 130:2111–2117
Ma JF, Yamaji N, Tamai K, Mitani N (2007) Genotypic difference in silicon uptake and expression of silicon transporter genes in rice. Plant Physiol 145:919–924
Marschner H (1995) Beneficial mineral elements. In: Mineral nutrition of higher plants, 2nd edn. Academic Press, San Diego, California, pp 405–435
Marshall WE (2004) Utilization of rice hull and rice straw as adsorbents. In: Champagne ET (ed) Rice chemistry and technology. American Association of Cereal Chemists, Inc., St. Paul, pp 611–630
Mitani N, Ma JF, Iwashita T (2005) Identification of the silicon form in xylem sap of rice (Oryza sativa L.). Plant Cell Physiol 46:279–283
Mitani N, Yamaji N, Ma JF (2008) Characterization of substrate specificity of a rice silicon transporter, Lsi1. Eur J Physiol 456:679–686
Nei M (1973) The theory and estimation of genetic distance. In: Morton NE (ed) Genetic structure of populations. University Press of Hawaii, Honolulu, pp 45–54
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Prabbu AS, Barbosa Filho MP, Filippi MC, Datnoff LE, Snyder GH (2001) Silicon from rice disease control perspective in Brazil. In: Datnoff LE, Snyder GH, Korndorfer GH (eds) Studies in plant science 8: silicon in agriculture. Elsevier Science B.V, Amsterdam, pp 293–311
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Proctor A (1990) X-ray diffraction and scanning electron microscope studies of rice hull ash. J Am Oil Chem Soc 67:576–584
Rodrigues FÁ, McNally DJ, Datnoff LE, Jones JB, Labbé C, Benhamou N, Menzies JG, Bélanger RR (2004) Silicon enhances the accumulation of diterpenoid phytoalexins in rice: a potential mechanism for blast resistance. Phytopathology 94:177–183
Saito K, Yamamoto A, Sa T, Saigusa M (2005) Rapid, micro-methods to estimate plant silicon content by dilute hydrofluoric acid extraction and spectrometric molybdenum method: I. Silicon in rice plants and molybdenum yellow method. Soil Sci Plant Nutr 51:29–36
SAS Institute Inc., (1999) SAS Users Guide, Version 8.02. Statistical Analysis System Institute, Cary, North Carolina
SAS Institute Inc., (2008) SAS/STAT 9.2 User’s Guide. SAS Institute Inc, Cary, NC
Singh M, Ceccarelli S, Hamblin J (1993) Estimation of heritability from varietal trials data. Theor Appl Genet 86:437–441
Sneller CH, Mather DE, Crepieux S (2009) Analytical approaches and population types for finding and utilizing QTL in complex plant populations. Crop Sci 49:363–380
Sun L, Gong K (2001) Silicon-based materials from rice husks and their applications. Ind Eng Chem Res 40:5861–5877
Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18:283–294
Takahashi E (1995) Uptake mode and physiological functions of silica. In: Matsuo T, Kumazawa K, Ishii U, Ishihara K, Hirata H (eds) Science of the rice plant, vol 2 physiology, vol 2. Food and Agricultural Policy Research Center, Tokyo, pp 420–433
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wang X, Fjellstrom R, Jia Y, Yan WG, Jia MH, Scheffler BE, Wu D, Shu Q, McClung A (2010) Characterization of Pi-ta blast resistance gene in an international rice core collection. Plant Breed 129:491–501
Wu Q-S, Wan X-Y, Su N, Cheng Z-J, Wang J-K, Lei C-L, Zhang X, Jiang L, Ma J-F, Wan J-M (2006) Genetic dissection of silicon uptake ability in rice (Oryza sativa L.). Plant Sci 171:441–448
Xin Z, Velten JP, Oliver MJ, Burke JJ (2003) High-throughput DNA extraction method suitable for PCR. Biotechniques 34:802–826
Yamaji N, Mitatni N, Ma JF (2008) A transporter regulating silicon distribution in rice shoots. Plant Cell 20:1381–1389
Yan W, McClung A (2010) ‘Rondo’, a long-grain indica rice with resistances to multiple diseases. J Plant Regist 4:131–136
Yan W, Rutger JN, Bryant RJ, Bockelman HE, Fjellstrom RG, Chen M-H, Tai TH, McClung AM (2007) Development and evaluation of a core subset of the USDA rice germplasm collection. Crop Sci 47:869–878
Yu J, Pressoir G, Briggs WH, Vroh BL, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed model for association mapping that accounts for multiple levels of relatedness. Nat Genet 38(2):203–208
Yu J, Zhang Z, Zhu C, Tabanao D, Pressoir G, Tuinstra M, Kresovich S, Todhunter R, Buckler E (2009) Simulation appraisal of the adequacy of number of background markers for relationship estimation in association mapping. Plant Genome 2:63–77
Zhang J, Zhu YG, Zeng DL, Cheng WD, Qian Q, Duan GL (2007) Mapping quantitative trait loci associated with arsenic accumulation in rice (Oryza sativa). New Phytol 177:350–355
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Acknowledgments
The authors thank J. Neil Rutger (Research Geneticist, retired) at the Dale Bumpers National Rice Research Center for providing seeds of Wells brittle, Heather Howe for technical assistance in silica concentration analysis, Tiffany Sooskaserm, Yao Zhou, Biaolin Hu, Yuemei Huang, Jodie Cammack, Kip Landry, Carl Henry, Jason Bonnette, Piper Roberts, LaDuska Simpson, and Sarah Hendrix for assistance in conducting the field studies, and Melissa Jia for genotyping. The XRD results were generated at the University of Arkansas using the Electron Optics Facility, partly funded by the NSF.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mention of a trademark or proprietary product in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture. USDA is an equal opportunity provider and employer.
Rights and permissions
About this article
Cite this article
Bryant, R., Proctor, A., Hawkridge, M. et al. Genetic variation and association mapping of silica concentration in rice hulls using a germplasm collection. Genetica 139, 1383–1398 (2011). https://doi.org/10.1007/s10709-012-9637-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-012-9637-x