Abstract
Doublesex (dsx) is a double-switch gene at the bottom of the somatic sex-determination hierarchy which regulates sexual dimorphism in many insects. Here, Drosophila melanogaster homologues of dsx were isolated in two Bactrocera species, the oriental fruit fly, B. dorsalis, and the guava fruit fly, B. correcta. Results of RT–PCR analysis suggests that both the B. dorsalis dsx (Bd1dsx) and B. correcta dsx (Bcdsx) genes are transcribed and sex-specifically spliced in accordance with the Drosophila sex-specific splicing mechanism. The cDNA sequences shared a high degree of similarity at the nucleotide level among the Bactrocera species. Structurally conserved domains for DNA-binding and oligomerization were observed in all transcripts suggesting that their proteins function as transcriptional factors for downstream sex-specific gene expression. A purine-rich element (PRE) and four repeat elements (dsxRE) for TRA/TRA-2 binding sites were also found in the 3′ untranslated regions (UTR) of both the female Bd1dsx and Bcdsx mRNAs. Notably, a putative core promoter was revealed in Bd1dsx, being probably the first dsx promoter discovered in the tephritid flies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Two true fruit flies species, B. dorsalis (Hendel), or oriental fruit fly, and B. correcta (Bezzi), or guava fruit fly, are commonly found in Southeast Asia, especially where commercial crops are grown. Thus, they present very serious pest insects for the fruit and vegetable markets both locally and internationally.
Preventive measures have been practiced widely in an attempt to reduce economical loss due to fruit fly prevention methods such as chemical use, methyl eugenol/protein baiting, fruit wrapping, as well as sterile insect technique (SIT). The latter method, particularly, seems to have advantages over the others in that SIT offers an attractive alternative fruit fly population control method that is species-specific, non-polluting, and safe to both humans and the environment. In most cases, only the sterile male flies are desired for release since the females, though sterile, can still damage the crops through oviposition, distract the male flies from mating with the wild females, transmit diseases, and increased costs of production and distribution. To come up with a more effective SIT program, a large-scale sex separation process using genetic sexing stains (GSSs) and competitive mating of sterile male flies are to be taken into consideration (Wimmer 2005). The masculinization of XX individuals using the tra—RNA interference technology is one of the most promising approaches in the generation of phenotypically male-only GSS (Pane et al. 2002; Lagos et al. 2007). Nonetheless, the true success of such a genetic manipulation is based on a better understanding of the genes involved in the sex determination pathways of the particular pests.
Somatic sex determination in D. melanogaster begins when a ratio of X chromosome: autosome (X:A) signals the RNA splicing cascade involving alternative splicing of Sex-lethal (Sxl), Transformer (tra), fru, and dsx genes, respectively. Flies with the X:A ratio of 1 develop into females while those with ratio of 0.5 grow into males (Cline 1993). Sxl is turned on only in the females and SXL protein maintains its own autoregulatory loop while regulating a productive splicing of a downstream gene tra (Boggs et al. 1987; Inoue et al. 1990). Together with non-sex specifically expressed TRA-2 protein, TRA activates a female-specific splicing of the dsx gene, the gene at the bottom of sex-determination hierarchy, resulting in DSXF protein (Hodgkin 1989). In contrast, once the Sxl gene is turned off in the males, the genes in the rest of the cascade splice in a male-specific manner and lead to a default mode of splicing the dsx mRNA yielding DSXM protein. Both proteins resulting from different modes of splicing of the dsx mRNA are functional but opposite to each other as transcriptional factors of the genes downstream controlling sexual dimorphism (Burtis and Baker 1989). However, sex determination in the tephritid insects probably differs in the initial signal and/or the master switch gene. Even though a highly-conserved homologue exists, Sxl is equally expressed in both male and female Ceratitis capitata (medfly). CcSxl gene has no sex-specific variants, so its expression cannot affect the sex fate (Saccone et al. 1998). Instead, Cctra, along with its positive autoregulatory loop and a non-sex specific Cctra-2 gene, was proposed to be a key master gene for female sex determination of the medfly (Pane et al. 2002; Salvemini et al. 2009). It was shown that RNAi targeting tra-2 aux-ep directly and tra indirectly led to a simultaneous change in the sex-specific splicing of dsx and fru (Salvemini et al. 2009). Outside the drosophilids, Sxl, tra, tra-2, dsx, and fru homologues were identified in many tephritid insects and their high degree of conservation, especially regarding the dsx gene, suggests a preservation mechanism of these genes and a similar flow of information to that of the Drosophila tra/tra-2 > dsx/fru cascade (Schutt and Nothiger 2000; Saccone et al. 2002; Shearman 2002; Graham et al. 2003; Sanchez 2008).
At a very early stage of embryogenesis, sex determination is one of the main events in the Mother-to-Zygotic Transition (MZT) (Gouw et al. 2009; Gabrieli et al. 2010). Consequently, a precise and careful reprogramming of maternally-inherited transcripts of the sex determination genes is needed in order to establish sex in the flies. Gabrieli et al. (2010) hypothesized that maternal information of embryonic development is reset via Cctra mRNA splicing and a degradation of maternally-inherited Ccdsx transcripts. An XX embryo develops female-specific characteristics mainly through the positive autoregulatory loop of Cctra despite a degradation of maternally-inherited transcripts. For an XY embryo to develop its male characteristics, the M factor located on the Y chromosome might have an effect on mRNA splicing or protein activity which leads to an inhibition of the female-specific autoregulatory loop of tra and results in the male mode of Cctra splicing (Willhoeft and Franz 1996; Gabrieli et al. 2010). Additionally, the presence of sex-specifically spliced transcripts of Ccdsx was reported to begin in 10 h embryos. Therefore, the sex determination cascade in the medfly is assumed to be completed before the end of the cellular blastoderm formation, because cellularization in C. capitata starts later than that of D. melanogaster, and the medfly’s sex determination cascade is shorter (Gabrieli et al. 2010). Furthermore, a recent finding in D. melanogaster addressed a major revision on how sex-specific function is regulated in flies. In addition to a single regulatory event of an RNA-splicing cascade, elaborate temporal and spatial transcriptional controls of the terminal genes, dsx and fru are also involved in sexual differentiation of paticular tissues during the early development (Robinett et al. 2010).
Being one of the final regulatory genes in the insect sex determination pathway, the dsx gene has been characterized in many dipterans such as Anopheles gambiae (Scali et al. 2005), Musca domestica (Hediger et al. 2004), Megaselia scalaris (Sievert et al. 1997; Kuhn et al. 2000), in the lepidopteran Bombyx mori (Ohbayashi et al. 2001; Suzuki et al. 2001), in the hymenopteran Apis mellifera (Cho et al. 2007), and in the fruit flies B. tryoni (Queensland fruit fly) (Shearman and Frommer 1998), B. oleae (olive fruit fly) (Lagos et al. 2005), B. dorsalis (oriental fruit fly) (Chen et al. 2008), C. capitata (medfly) (Saccone et al. 1996), and twelve species of Anastrepha (Ruiz et al. 2005; Ruiz et al. 2007). Other than that, many dsx functional studies have been performed to unveil the state of the art of dsx evolution. A knock-down experiment in female B. dorsalis adults with female-specific dsx dsRNA resulted in an interruption of yolk protein (yp) expression which led to a significant reduction in ovary size and number of oocytes as well as an abnormal formation of the reproductive organs (Chen et al. 2008). Other non-drosophilid insects whose dsx functional studies are available include Bombyx mori (silkworm) (Suzuki et al. 2003), M. domestica (Hediger et al. 2004), C. capitata (Saccone et al. 2008), and A. obliqua (Alvarez et al. 2009).
Apparently, dsx is the most highly conserved gene in the sex determination pathway (Permpoon and Thanaphum 2010). As well, having a shorter sex determination pathway and a slow rate of early developmental process in C. capitata (Gabrieli et al. 2010) and having a specific time and place of dsx expression in D. melanogaster (Robinett et al. 2010) make the dsx gene and its promoter available as an alternative tool in the study of the expression and splicing mechanisms involved in the MZT in Diptera as well as a genetic tool for population control against pest insects. Such advantages lead to this research which was to isolate and characterize the homologues of the dsx gene of the two economically important fruit flies in the Asia–Pacific regions. Note that two oligomerization domains of the B. dorsalis (Bd1dsx) and B. correcta (Bcdsx) dsx gene coding regions were briefly discussed in short communication by Permpoon and Thanaphum (2010). Therefore, the present study deals with the isolation and characterization of sex-determining dsx orthologues in B. dorsalis and B. correcta and their putative promoters. Our results showed that both of the genes are highly conserved in structure and function, analogous to those in other non-drosophilid insect species studied. After the full-length dsx cDNAs were obtained, RT–PCR with appropriate primers was carried out to confirm the sex-specific splicing patterns in the male and female fruit flies. Further, putative core promoter regions of the dsx gene were suggested in B. dorsalis and B. correcta, representing the first finding within the tephritid fruit flies.
Materials and methods
DNA and RNA extractions
Genomic DNA was extracted from laboratory stocks of adult B. dorsalis (Hendel) (Phyathai 1 strain) and adult B. correcta (Bezzi) (Phyathai 2 strain) essentially as described by Baruffi et al. (1995). The total RNA was isolated from laboratory stocks of adult B. dorsalis (Hendel) (Phyathai 1 strain) and adult B. correcta (Bezzi) (Phyathai 2 strain) by using Trizol reagent (Gibco/BRL Life Technologies, Gaithersburg, MD, USA) according to the manufacturer’s instructions.
3′ and 5′ cDNA RACE
3′ and 5′ cDNA RACE (Rapid Amplification of cDNA Ends) reactions were carried out essentially as described by Frohman et al. (1988). The ImProm-II reverse transcription system (Promega, Madison, WI, USA) with either an oligo(dT) adapter primer (3′ RACE) or a dsx-specific primer (5′ RACE) was used to reverse transcribe ~3–5 μg total RNA from adult flies in a 20 μl total volume as recommended by the manufacturer. Some of the 3′ RACE primers were designed according to the male dsx sequences of the following species: B. tryoni (Btdsx: AF029676), B. oleae (Bodsx: AJ547622), B. dorsalis (Bddsx: AY669317), and C. capitata (Ccdsx: AF434935). Other primers were previously used to isolate dsx genes from such species as B. tryoni (Shearman and Frommer 1998). However, 5′ RACE primers were primarily designed from the alignments of sequenced nucleotides from the 3′ RACE PCR, namely, Bd1dsx and Bcdsx (this work). Refer to Table 1 for the primer sequences used in this study.
All amplification reactions were performed using a FlexCycler PCR thermal cycler (Analytik Jena, Germany). After the first strand of cDNA was synthesized, one-tenth of the initial RT–PCR volume was used as a template in 3′ and 5′ cDNA RACE using Taq polymerase (Vivantis Technologies, Selangor, Malaysia).
Standard cycling conditions for 3′ RACE were as follows: 94°C 4 min, held at 72°C while Taq polymerase was added, then one cycle of 60–63°C 2 min and 72°C 2 min; 94°C 1 min, 60–63°C 2 min, 72°C 2 min, 5 cycles; 91°C 40 s, 58–60°C 2 min, 72°C 2 min, 28 cycles; one cycle of final extension at 72°C 7 min. The product of the first amplification reaction with the dsx-specific primer, BD, and the adapter primer (20 pmol) was used in the second-round amplification with the Btk primer. Third-round amplification was carried out using 2 μl of the previous round’s product as a template in a presence of the Btl primer.
Standard cycling conditions for 5′ RACE were as follows: 94°C 5 min; 94°C 1 min, 55–63°C 30 s, 72°C 2 min, 29 cycles; one cycle of final extension at 72°C 7 min. The reverse transcription product with RevBD primer was A-tailed using recombinant terminal deoxynucleotidyl transferase (rTdT) (Promega) according to the manufacturer’s specifications. The product from this reaction was then purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) before 2 μl was used as a template in an amplification with Btm_rev and adapter primers.
Inverse PCR
Inverse PCR was performed in order to find the 5′ upstream region of the dsx gene in both species. Five micrograms of genomic DNA was digested at 37°C using 50 units of CfoI (Promega) in a total volume of 100 μl. After 5 h, digested genomic DNA fragments were purified using the QIAquick PCR purification kit (QIAGEN). The purified digested DNA fragments were allocated to different quantities of 50, 100, and 200 ng and subsequently self-ligated in a volume of 100 μl at 14°C for 20 h. Purification of self-ligated DNA fragments (50, 100, and 200 ng) was done by ethanol precipitation. Primers were designed according to Bd1dsx and Bcdsx sequences (this work) and Shearman and Frommer (1998). Refer to Table 1 for the primer sequences used in this study.
Prior to an inverse PCR, a positive PCR was carried out with Bddn1 and Btl_rev primers following cycling conditions of: 94°C 2 min; 94°C 1 min, 60°C 30 s, 72°C 1 min, 30 cycles; one cycle of final extension at 72°C 7 min.
Inverse PCR was performed on the circularized fragments by using primer sequences in inverse orientation to the previously described positive PCR primers within the known dsx sequence of 5′ UTR. A PCR amplification was performed with Bddn1_rev and Btl primers following cycling conditions of: 94°C 7 min; 94°C 1 min, 60°C (depending on primers) 30 s, 72°C 5 min, 30 cycles; one cycle of final extension at 72°C 10 min. PCR product size was analyzed by agarose gel electrophoresis in comparison with λ HindIII-EcoRI marker and 100 bp DNA ladder marker (Promega). For verification of inverse PCR products, a nested PCR with Bdup1 and BDR primers was carried out following the same positive PCR profile.
Fragment isolation, cloning, and sequencing
PCR products were excised from 1% agarose gel and purified using the Geneclean II kit (Bio 101 Inc., La Jolla, CA, USA) and then ligated into the pGEM-T Easy vector (Promega) according to the manufacturers’ instructions. Recombinant plasmids were cloned into DH5α competent cells and isolated as described by Sambrook et al. (1989). All sequencing was performed on both strands using the ABI3730XL sequencing machine by Macrogen Inc., Seoul, Korea.
RT–PCR analysis
First strand cDNA of B. dorsalis and B. correcta were generated by the reverse transcription method as previously described using sex- and gene-specific primers. One-tenth of the initial RT–PCR volume was used in a standard PCR amplification using common and sex-specific dsx primers, following cycling conditions of: 94°C 2 min; 94°C 1 min, 55–62°C 30 s, 72°C 1 min, 29 cycles; one cycle of final extension at 72°C 7 min. Primers were designed according to Bd1dsx and Bcdsx sequences (this work) and Shearman and Frommer (1998). Refer to Table 1 for the primer sequences used in this study.
Sequence alignment and phylogenetic tree reconstruction
ClustalW (1.83) (Thompson et al. 1994) was used to align DNA and protein sequences. Phylogenetic trees were reconstructed based on genetic distance; 1,000 replications of bootstrapping and consensus phylogenetic trees with bootstrap values were drawn based on the unweighted pair-group method with arithmetic mean (UPGMA) using the CLC Main Workbench 4.0.1 package (CLC Bio, Aarhus, Denmark).
Results
Isolation of dsx homologues in B. dorsalis and B. correcta
In an attempt to acquire the cDNA fragments containing dsx coding regions, 3′ RACE, 5′ RACE, and RT–PCR techniques were employed. The expected products were amplified successfully using the newly designed primers and the specific, non-degenerate primers designed from the sequences of dsx orthologues from other tephritids.
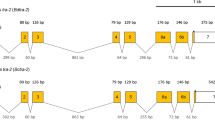
Sex-specific transcripts of Thailand’s B. dorsalis dsx gene, Bd1dsx, isolated from the male flies were ~2.9 kb long (Bd1dsx m: acc. no. FJ185162) and ~1.7 kb long from the female flies (Bd1dsx f: acc. no. FJ176944) (Fig. 1a). The complete coding sequences (CDS) were obtained: 1,203 bp open reading frame (ORF) coding for 400 amino acid residues in Bd1dsx m and 966 bp ORF coding for 321 residues in Bd1dsx f. The Bd1dsx CDS nucleotide and deduced amino acid sequences were consistent with those of previously isolated B. dorsalis native to the island of Taiwan (Chen et al. 2008) and showed high similarities among the tephritid fruit flies.
Nucleotide sequences of male and female cDNAs and 5′-flanking genomic DNA, and predicted amino acid sequences of the male and female dsx polypeptides belonging to Bd1dsx (a) and Bcdsx (b). Nucleotides are numbered in the right margin from the beginning of the presented sequence. At the end of the last common exon, numbering continues independently for the female- and male-specific exons, with coordinates in the sex-specific sequences designated by the superscripts “f” and “m”. The sequences encoding the major open reading frame are separated into common, female-specific, and male-specific regions, and the amino acids are numbered in the right margin in a manner analogous to that used for the nucleotides. In the 5′-flanking region, blue-shaded putative CAAT boxes (Bd1dsx: nucleotides 1,282–1,285, Bcdsx: nucleotides 397–400), pink-shaded TATA boxes (Bd1dsx: nucleotides 1,328–1,335, Bcdsx: nucleotides 407–414) and violet-shaded initiator sequences (Bd1dsx: nucleotides 1,357–1,363, Bcdsx: nucleotides 424–430) are illustrated. Female-specific dsxREs and PREs are highlighted in gray and yellow. The IX binding regions are underlined in red (Bd1DSX: amino acids 244–302, BcDSX: amino acids 244–302). Polyadenylation signals in both female- and male- specific sequences are marked with the green boxes. Sequence data have been submitted to the GenBank data library: accession numbers FJ176944 for Bd1dsx f, FJ185162 for Bd1dsx m, FJ185166 for Bcdsx f, and FJ185165 for Bcdsx m
The Guava fruit fly’s complete CDS of the dsx gene, Bcdsx, was also recovered in both sexes. A ~2.4 kb male transcript of Bcdsx (Bcdsx m: acc. no. FJ185165) contained 1,203 bp ORF, coding for 400 amino acids and a ~1.9 kb female transcript (Bcdsx f: acc. no. FJ185166) had 966 bp ORF, coding for 321 amino acids (Fig. 1b). Both male- and female- Bcdsx transcripts were identical in the numbers of nucleotide and deduced amino acids to those of the oriental fruit fly’s Bd1dsx gene.
Conservation of the dsx gene across the Bactrocera genus
Doublesex transcripts isolated from B. dorsalis of Thai and Taiwanese origins were almost identical. Bd1dsxm had 99% nucleotide and 100% amino acid identities to the Taiwanese Bddsxm, and Bd1dsxf had 99% similarity at both nucleotide and amino acid levels to the Taiwanese Bddsxf. Moreover, the identity at the nucleotide level of the Thai oriental fruit fly’s Bd1dsx was 95–97% within the Bactrocera group: B. tryoni (Btdsx: Shearman and Frommer 1998), B. oleae (Bodsx: Lagos et al. 2005) and B. correcta (Bcdsx: this work) whereas a lower range of 82–85% identity was observed in a more distantly-related species within the same Tephritidae family as in A. obliqua (Aodsx: Ruiz et al. 2005) and C. capitata (Ccdsx: Saccone et al., unpublished, acc. no.’s AF434935 and AF435087). Accordingly, 97–98 and 89–93% similarities at the predicted amino acid level were perceived among a Bactrocera group and a non-Bactrocera group (A. obliqua and C. capitata), respectively.
Similarly, Bcdsx had an identity at a CDS nucleotide level of 95–97% within the species of the same genus and decreased to 82–85% in a non-Bactrocera group. The similarity at an amino acid level was 97–98 and 88–92% in the Bactrocera and non-Bactrocera groups, respectively. The percentage of nucleotide identity and amino acid similarity of the guava fruit fly followed the same trend observed in the oriental fruit fly, suggesting that these dsx transcripts were conserved and, most likely, still had functions in the sex-determination pathway of the fruit fly. Moreover, the results of the phylogeny tree were in conformity with a Clustal alignment of dsx CDS. Figure 2 illustrates a close relationship within the Bactrocera genus and its distinct separation from the other groups, especially from the drosophilid family.
Molecular phylogenies reconstructed from the female (a) and male (b) DNA sequences of the coding region of the dsx gene. Both trees were reconstructed using the UPGMA method. The horizontal branch-lengths are proportional to the genetic distance and the numbers shown at branch points indicate bootstrap values from 1,000 replicates
The putative DSX proteins of B. dorsalis and B. correcta
In B. dorsalis, the ORF of female the dsx transcript coded for a putative female-specific protein, Bd1DSXF, contained 321 amino acids. The first set of codon, ATG, was located in the 5′ common segment (at position 141 in acc. no. FJ176944) and the ORF ended in the female-specific region (TGA at position 1,106). The longer ORF in the male-specific transcript coded for a putative male-specific protein, DSXM, was 400 amino acids in length. The ORF started at the same site as in the female ORF in the 5′ common region (ATG at position 793 in acc. no. FJ185162) and ended with a TAA at position 1995 in a male-specific region. Similarly, the ORF in B. correcta females began in the 5′ common fragment (at position 309 in acc. no. FJ185166) and ended in the female-specific region with TGA at position 1,274, coding for a 321 putative amino acid-long BcDSXF protein. Correspondingly, the male ORF coded for 400 putative amino acids of BcDSXM which started at the same site in the 5′ common region (ATG at position 313 in acc. no. FJ185165) and ended in the male-specific segment with TAA at position 1,515.
Further analysis of the Bd1DSX and BcDSX proteins showed that both male- and female- amino acid sequences shared a common N-terminal region but differed at the sex-specific C-terminal region. The shared common amino terminus covered the first 291 amino acid residues (Fig. 3a) and contained a zinc finger-like DNA-binding domain (OD1). OD1 plays a vital role in DNA binding and protein oligomerization in the Drosophila DSX protein (An et al. 1996), and its homologue was located between amino acids 39–104 of both Bd1DSX and BcDSX. Another dsx molecular feature that both sexes shared was an oligomerization domain (OD2). OD2 is required for an oligomerization of Drosophila DSX and it consists of sex-specific and non-sex specific sequences. The non-sex specific part of OD2 covered amino acids 244–291 of both sexes while the sex-specific part comprised amino acids 292–306 (15 residues) in the females and amino acids 292–327 (36 residues) in the males (Fig. 3b, c).
Comparison of the DSX predicted polypeptides in B. dorsalis (Bd) (this work), B. correcta (Bc) (this work), B. tryoni (Bt) (Shearman and Frommer 1998), B. oleae (Bo) (Lagos et al. 2005), C. capitata (Cc) (Saccone, unpublished data 2001), and A. obliqua (Ao) (Ruiz et al. 2005). a Sequence common to both sexes, b female-specific sequence and c male-specific sequence. The DNA-binding domain OD1 and the oligomerization domain OD2 are boxed in dashed and solid lines, respectively. Gaps were introduced in the alignments to maximize similarity. The comparison of protein sequences was performed using ClustalW (1.83)
Next, the sex-specific C-termini of DSX were examined (amino acids 292–321 in females and 292–400 in males). The female-specific C-termini of the DSX protein in both species (30 amino acids) were shorter than those of the males (109 amino acids). The difference in size of the C-termini between the two sexes was in agreement with patterns of DSXF and DSXM found in D. melanogaster, B. tryoni, and B. oleae (Burtis and Baker 1989; Shearman and Frommer 1998; Lagos et al. 2005). Moreover, a conserved putative binding region of intersex (IX), an obligatory partner protein and putative transcriptional coactivator of Drosophila DSXF (Yang et al. 2008), was also identified here in Bd1DSX and BcDSX in a span of 59 amino acids (Fig. 1a, b).
A BLASTX search with Bd1DSXF (acc. no. FJ176944) and BcDSXF (acc. no. FJ185166) in the non-redundant (nr) sequence database of NCBI returned the BdDSXF, BoDSXF and BtDSXF entries from B. dorsalis, B. oleae, and B. tryoni, respectively, with the highest scores (93–99% identities) while the AoDSXF and CcDSXF entries of A. obliqua and C. capitata held 90–91% identities to the query sequences. Similar results were obtained when a BLASTX search of Bd1DSXM (acc. no. FJ185162) and BcDSXM (acc. no. FJ185165) was performed.
Regulatory elements in female-specific exons of Bd1dsx and Bcdsx
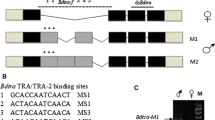
Four homologues of the 13-nucleotide repeat sequence (dsxRE) and the homologues of purine-rich enhancer (PRE) sequences were found in the 3′ UTR of Bd1dsx f and Bcdsx f (Fig. 4a, b). There were poly(A) signals near the 3′ end of female-specific transcripts which also appeared in Bodsx of B. oleae (Lagos et al. 2005). A substantial similarity to the Drosophila dsxRE and PRE can be seen in Tables 2 and 3. Note that higher similarity of the two elements was observed among the fruit flies in the Bactrocera genus. The presence of dsxRE/PRE clusters in Bd1dsx f and Bcdsx f and their relative conservation at a nucleotide level among the genus suggest that a sex-specific splicing mechanism of the pre-mRNA through an activation of selected female-exon similar to that in Drosophila might also take place in the sex determination pathway of oriental and guava fruit flies.
The female-specific exons of B. dorsalis dsx (a) and B. correcta dsx (b).Translational stop codon (TGA) and polyadenylation signals are shaded in red and green, respectively. Distribution of the 13 nucleotides repeats and the PRE are also highlighted in the sequence in gray and yellow boxes, respectively
RT–PCR analysis of sex-specific splicing in Bd1dsx and Bcdsx
In order to determine whether a sex-specific splicing took place in Bd1dsx and Bcdsx, RT–PCR with appropriate primers was carried out. Sex-specific first strand cDNA of both species under study was generated using sex- and gene-specific primers whose binding sites are located in the 3′ UTR of the dsx gene. Three sets of RT–PCR reactions were carried out in the males and females with primers designed to amplify the common region and female- and male-specific portions of the dsx transcript (see Materials and methods for list of primers). Figure 5 depicts the location of primer-pairs and detection of the sex-specific Bd1dsx and Bcdsx transcripts by RT–PCR. The common region of the dsx transcript was successfully amplified in both sexes of B. dorsalis and B. correcta. As expected, no product was detected on the electrophoresis gel when female cDNA was used for amplification with a primer-pair designed for male-specific region and vice versa when male cDNA was used for amplification with female-specific primers. The unexpected bands of ≤100 bp amplification products observed in lanes 3, 5, and 6 in Fig. 5b can be explained as a potential primer-dimer phenomenon. In agreement with other tephritid species previously determined, such results from oriental and guava fruit flies imply that a sex-specific splicing mechanics is located in their dsx genes as well.
Detection of sex-specific Bd1dsx and Bcdsx transcripts by RT–PCR analysis. The total RNA was prepared from adult male and female flies: B. dorsalis (a) and B. correcta (b). PCR was performed using primer dsx c1-c2 in lanes 1–4, primers dsx c3-f in lanes 5–7, and primers dsx c4-m in lanes 8–10. The location of the primers on cDNA clones is shown above. Positive control was performed on genomic DNA as a template (lane 4) and lanes 3, 7, and 10 are without RT. MW is 100 base pairs molecular weight DNA marker that steps from 100 to 1,000 base pairs in 100 base pairs increment
Inverse PCR to locate the putative core promoters
Once a complete CDS of Bd1dsx and Bcdsx was obtained and sequenced, primers were designed to amplify the genomic sequence flanking the 5′ UTR in pursuit of a core promoter regulatory region. In the case of B. dorsalis, inverse PCR analysis from genomic DNA templates revealed a low degree of nucleotide sequence conservation at the 5′ end flanking a short stretch (100 bp) of highly conserved sequence (99%) that may represent a putative core promoter (data not shown). Analysis of the 2,330 bp and 767 bp upstream region of the dsx gene in B. dorsalis (acc. no. FJ185163) and B. correcta (acc. no. FJ185164), respectively, revealed TATA boxes and several other consensus RNA Polymerase II transcriptional factor recognition sequences offering evidence for a putative core promoter region. The putative core promoter regulatory region in Bd1dsx was composed of the CAAT box, TATA box, and initiator (Inr) sequence covering positions −1,049 through −968 upstream of the Bd1dsx start codon (Fig. 1a). Also made up of the same three recognition elements, the putative core promoter regulatory region in Bcdsx spanned positions −371 through −338 upstream of the start codon (Fig. 1b). The TATA box of Bd1dsx matched the consensus sequence TATAWAAR (W is A or T/R is A or G) but that of Bcdsx had one mismatch (a G instead of an A). Each of the Inr sequences of both species had one deviation at the third position (G instead of A) in the Inr consensus sequence CC/TT AN TCC/ATT. A preference regarding the start site of the TATA box was featured in the Bd1dsx putative core promoter region in that the upstream ‘T’ in the TATA box is located at position −31 relative to the ‘A’ in the Inr consensus sequence (in this case a ‘G’), common to most TATA boxes and Inr locations (Butler and Kadonaga 2002; Juven-Gershon et al. 2006). Similar regulatory elements of the putative core promoter were also recognized in Bcdsx. The presence of the putative CAAT box, TATA box and Inr sequence suggests the possibility of a core promoter because, consistently, similar features of these core promoters were observed in Bodsx of B. oleae as well (Lagos et al. 2005). This may be the first time that these core promoters have been recognized in the dsx gene of insects.
Discussion
Conservation of structure and function in Bd1dsx and Bcdsx
Bd1dsx and Bcdsx were present and expressed in a sex-specific manner in adult male and female flies of both B. dorsalis and B. correcta. The molecular organization of Bd1dsx and Bcdsx was similar to that of the model organism D. melanogaster: the female and male mRNAs shared the first three exons and differed in the remaining downstream exons. Hence, the female transcript distinctively comprised the exons 1–3 homologous sequence with the addition of the female-specific exon 4 homologue. On the other hand, the male transcript consisted of the first three common exon’s homologous sequence plus the male-specific exons 5 and 6 homologues. The alternative processing of mRNA of the same gene appeared to be the mode of sex-specific transcript production of dsx in B. dorsalis and B. correcta rather than the sex-specific expression of two different genes. Common region and sex-specific primer pairs can successfully and equally amplify the correct fragments of expected size from the extracted genomic DNA of both sexes (data not shown). In contrast, only common fragments can be amplified from both male and female cDNA pools. No amplification product was detected with female cDNA and male-specific primer pairs and vice versa. Additionally, the assembled sequences of Bd1dsx and Bcdsx transcripts showed that they shared a common 5′ region followed by an alternative 3′ region sequence in the female- and male-specific transcripts. Consequently, these sex-specific transcripts would encode for female-specific, DSXF, and male-specific, DSXM, proteins sharing the common N-terminal region but differ in their sex-specific C-terminal domain (Burtis and Baker 1989). Bd1dsx and Bcdsx contained OD1, a zinc-finger domain involved in DNA binding and oligomerization, and OD2, a domain required for the oligomerization of the DSX protein consisting of sex-specific and non-sex specific sequences (An et al. 1996; Cho and Wensink 1997, 1998; Permpoon and Thanaphum 2010). The amino acid sequences of the two domains of DSXF proteins of the oriental fruit fly and guava fruit fly revealed 95–100% similarity to those of B. oleae, B. tryoni, A. obliqua, C. capitata, and to a lesser extent (87–93%) those of D. melanogaster. The high similarity in the deduced amino acid sequences of Bd1dsx and Bcdsx is consistent with a common evolutionary origin of the dsx gene in fruit flies and its conserved function as the transcriptional regulator governing the downstream somatic sexual differentiation genes in both sexes (Burtis and Baker 1989). As well, the homology of the DNA binding structure (DM motif) was also discovered in C. elegans and, later on, in vertebrates such as mice, chickens, and humans (Raymond et al. 1998, 1999a, b). Thus, this conservation at the amino acid level of dsx, a teminal gene of the sex determination cascade, provided supporting evidence to endorse the theory that the sex-determining hierarchy in a variety of organisms ranging from insects to mammals evolved from the bottom to the top (Wilkins 1995). The importance of dsx as a double-switch key regulator in the sex-determination pathway was highlighted here by means of domain conservation. Furthermore, a successful manipulation of dsx gene in one species should be, at least in principle, transferrable to other species of the same genus without much difficulty as seen in the successful manipulation of the Cctra to develop the medfly sexing strains. The construct of a female-specific autocidal genetic system can even work in the Drosophila transgenic flies, underlying the potential transferability of the genetic sexing strategy in any ditperan species (Fu et al. 2007).
Regulatory elements conservation in female-specific dsx transcripts
The mechanism of sex-specific splicing regulation found in Drosophila appeared to play a similar role in B. dorsalis and B. correcta as a comparison of putative male and female amino acid transcripts revealed that the female-specific exon (exon 4) was skipped over in the males, and that the female dsx transcripts harbored four putative TRA/TRA-2 binding dsxRE and PRE as found in the dsx f transcripts of D. melanogaster, B. tryoni, B. oleae, and A. obliqua. Since tra genes have been identified in C. capitata (Pane et al. 2002), B. oleae (Lagos et al. 2005), and a number of fruit flies in the Anastrepha genus (Ruiz et al. 2007), it was suspected that B. dorsalis and B. correcta might also have the orthologous tra gene and that its product would regulate the dsx splicing in a maner comparable to that of D. melanogaster. In addition, a binding region of IX, a partner protein and putative transcriptional coactivator of DSXF in Drosophila, was also identified in both female-specific dsx transcripts (see red underline in Fig. 1a, b). Yang et al. (2008) has described the solution structure of the C-terminal domain of D. melanogaster DSXF and its functional implications. The binding of IX is mediated by the proximal helical portion of the female tail which is composed of UBA-like alpha helices spanning nearly the entire OD2 domain. The importance of steric and electrostatic complementarity across the interface is emphasized by mutagenesis of this portion. Therefore, a high identity in the amino acid sequence at the IX binding site on exon 4 of Bd1dsx f and Bcdsx f implies that such interactions might also occur in oriental and guava fruit flies as well.
Discovery of putative core promoter regions in Bd1dsx and Bcdsx
A unique finding in this work was the discovery of the putative core promoter region in the dsx gene of the oriental fruit fly, B. dorsalis. Core promoter is defined as a site of action of RNA polymerase II transcriptional machinery comprised the TATA box, Inr, and TFIIB recognition element (BRE). Although core promoter elements are dynamic and vital participants in the regulation of transcriptional activity, it is important to keep in mind that each of these core promoter elements is found in some, but not all, core promoters and that a considerable diversity in core promoter structure and function has been revealed in recent studies. For instance, some promoters are TATA-less but instead contain multiple GC box motifs (reviewed in Butler and Kadonaga 2002). Similarly, certain characteristics are essential and required for the core promoter to function efficiently, such as the presence of the Inr sequence. Identified in the 5′ flanking genomic DNA sequence, the putative core promoter regulatory region of Bd1dsx m was adjacent to a putative initiation site. The putative core promoter region included three core promoter elements: the CAAT box, the TATA box, and the Inr sequence. However, the Inr sequence of Bd1dsx, TTGCATT, contained one mismatch from the consensus (CC/TT AN TCC/ATT) in that the third position is a ‘G’ instead of an ‘A’ (Watson et al. 2008). The location of the upstream ‘T’ in the TATA box was exactly at −31 position relative to the ‘A’ in the Inr consensus sequence (a ‘G’ in this case) concurring with the TATA box location preference with regard to the Inr motif (Juven-Gershon et al. 2006). The putative core promoter region in Bcdsx did not possess all features as Bd1dsx m did. Therefore, it was proposed that it functions as a weaker promoter with the presence of the TATA box and several RNA polymerase II recognition sequences.
As a terminal gene in the sex determination cascade, the dsx structural characterization and its expression uncovered in this current study agreed with information from other closely related species, for which some functional studies have been performed. It is also accepted as one of the most conserved genes in the sex determination hierarchy. Therefore, it can be inferred that the novel putative promoters discovered here might be highly conserved in other closely-related species. The novel identification of a putative promoter will be useful in the future to perform comparative studies of this region in other dsx orthologues in the tephritids as well as to identify the regulatory regions involved in its transcriptional control. In addition, these promoters may be functional at a very early developmental stage in various insect tissues (Gabrieli et al. 2010; Robinett et al. 2010). Thus, the novel putative promoters may be further characterized in order to develop a genetic switch for gene manipulations of sex determination among tephritid insects.
References
Alvarez M, Ruiz MF, Sanchez L (2009) Effect of the gene doublesex of Anastrepha on the somatic sexual development of Drosophila. PLoS ONE 4(4):e5141. doi:10.1371/journal.pone.0005141
An W, Cho S, Ishii H, Wensink PC (1996) Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol Cell Biol 16:3106–3111
Baruffi L, Damiani G, Guglielmino CR, Bandi C, Malacrida AR, Gasperi G (1995) Polymorphism within and between populations of Ceratitis capitata: comparison between RAPD and multilocus enzyme electrophoresis data. Heredity 74:425–437
Boggs RT, Gregor P, Idriss S, Belote JM, McKeown M (1987) Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50:739–747
Burtis KC, Baker BS (1989) Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56:997–1010
Butler JE, Kadonaga JT (2002) The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev 16:2583–2592
Chen S, Dai S, Lu K, Chang C (2008) Female-specific doublesex dsRNA interrupts yolk protein gene expression and reproductive ability in oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Biochem Mol Biol 38:155–165
Cho S, Wensink PC (1997) DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J Biol Chem 272:3185–3189
Cho S, Wensink PC (1998) Linkage between oligomerization and DNA binding in Drosophila doublesex proteins. Biochemistry 37:11301–11308
Cho S, Huang ZY, Zhang J (2007) Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177:1733–1741
Cline TW (1993) The Drosophila sex determination signal: how do flies count to two? Trends Genet 9:386–390
Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNAs from rare transcripts: amplifications using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85:8998–9002
Fu G, Condon KC, Epton MJ, Gong P, Jin L, Condon GC, Morrison NI, Dafa’alla TH, Alphey L (2007) Female-specific insect lethality engineered using alternative splicing. Nature Biotech 25:353–357
Gabrieli P, Falaguerra A, Siciliano P, Gomulski LM, Scolari F, Zacharopoulou FG, Malacrida AR, Gasperi G (2010) Sex and the single embryo: early development in the Mediterranean fruit fly, Ceratitis capitata. Dev Biol 10:12. doi:10.1186/1471-213X-10-12
Gouw JW, Pinkse MW, Vos HR, Moshkin Y, Verrijzer CP, Heck AJ, Krijgsveld J (2009) In vivo stable isotope labeling of fruit flies reveals post-transcriptional regulation in the maternal-to-zygotic transition. Mol Cell Proteomics 8(7):1566–1578
Graham P, Penn JKM, Schedl P (2003) Masters change, slaves remain. Bio Essays 25:1–4
Hediger M, Burghardt G, Siegenthaler C, Buser N, Hilfiker-Kleiner D, Duendorfer A, Bopp D (2004) Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev Genes Evol 214:29–42
Hodgkin J (1989) Drosophila sex determination: a cascade of regulated splicing. Cell 56:905–906
Inoue K, Hoshijima K, Sakamoto H, Shimura Y (1990) Binding of the Drosophila Sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature 344:461–463
Juven-Gershon T, Hsu JY, Kadonaga JT (2006) Perspectives on the RNA polymerase II core promoter. Biochem Soc Trans 34:1047–1050
Kuhn S, Sievert V, Traut W (2000) The sex-determining gene doublesex in the fly Megaselia scalaris: conserved structure and sex-specific splicing. Genome 43:1011–1020
Lagos D, Ruiz MF, Sanchez L (2005) Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determination Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 348:111–121
Lagos D, Koukidou M, Savakis C, Komitopoulou K (2007) The transformer gene in Bactrocera oleae: the genetic switch that determines its sex fate. Insect Mol Biol 16:221–230
Ohbayashi F, Suzuki M, Mita K, Okano K, Shimada T (2001) A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp Biochem Physiol 128:145–158
Pane A, Salvemini M, Delli BP, Polito C, Saccone G (2002) The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129:3715–3725
Permpoon R, Thanaphum S (2010) Isolation and characterization of oligomerization domain I and II coding regions of doublesex genes in agricultural fruit flies (Diptera: Tephritidae). Eur J Entomol 107:121–126
Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (1998) Evidence for evolutionary conservation of sex-determining genes. Nature 391:691–695
Raymond CS, Parker ED, Kettlewell JR, Brown LG, Page DC, Kusz K, Jaruzelska J, Reinberg Y, Flejter WL, Bardwell VJ, Hirsch B, Zarkower D (1999a) A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum Mol Gen 8:989–996
Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D (1999b) Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol 215:208–220
Robinett CC, Vaughan AG, Knapp JM, Baker BS (2010) Sex and the single cell. II. There is a time and place for sex. PLoS Biol 8(5):e1000365. doi:10.1371/journal.pbio.1000365
Ruiz MF, Stefani RN, Mascarenhas RO, Perondini ALP, Selivon D, Sanchez L (2005) The gene doublesex of the fruit fly Anastrepha obliqua (Diptera, Tephritidae). Genetics 171:849–854
Ruiz MF, Eirin-Lopez M, Stefani RN, Perondini ALP, Selivon D, Sanchez L (2007) The gene doublesex of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. Dev Genes Evol 217:725–731
Saccone G, Peluso I, Testa G, Di Paola F, Pane A, Polito LC (1996) Drosophila Sex-lethal and doublesex homologous genes in Ceratitis capitata: searching for sex-specific genes to develop a medfly transgenic sexing strain, in enhancement of the sterile insect technique through genetic transformation using nuclear techniques. IAEA/FAO, Vienna
Saccone G, Peluso I, Astiaco D, Giordano E, Bopp D, Polito LC (1998) The Ceratitis capitata homologue of the Drosophila sex-determining gene Sex-lethal is structurally conserved, but not sex-specifically regulated. Development 125:1495–1500
Saccone G, Pane A, Polito LC (2002) Sex determination in flies, fruit flies and butterflies. Genetica 116:15–23
Saccone G, Salvemini M, Pane A, Polito LC (2008) Masculinization of XX Drosophila transgenic flies expressing the Ceratitis capitata DoublesexM isoform. Int J Dev Biol 52:1051–1057
Salvemini M, Robertson M, Aronson B, Atkinson P, Polito LC, Saccone G (2009) Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int J Dev Biol 53:109–120
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sanchez L (2008) Sex-determining mechanisms in insects. Int J Dev Biol 52:837–856
Scali C, Catteruccia F, Li Q, Crisanti A (2005) Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J Exp Biol 208:3701–3709
Schutt C, Nothiger R (2000) Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127:667–677
Shearman DCA (2002) The evolution of sex determination system in dipteran insects other than Drosophila. Genetica 116:25–43
Shearman DC, Frommer M (1998) The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol Biol 7:355–366
Sievert V, Kuhn S, Traut W (1997) Expression of the sex determination cascade genes Sex-lethal and doublesex in the phorid fly Megaselia scalaris. Genome 40:211–214
Suzuki MG, Ohbayashi F, Mita K, Shimada T (2001) The mechanism of sex specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem Mol Biol 31:1201–1211
Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T (2003) Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev Genes Evol 213:345–354
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R (2008) Molecular biology of the gene, 6th edn. Benjamin Cummings, California
Wilkins AS (1995) Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17:71–77
Willhoeft U, Franz G (1996) Identification of the sex-determining region of the Ceratitis capitata Y chromosome by deletion mapping. Genetics 144(2):737–745
Wimmer EA (2005) Eco-friendly insect management. Nat Biotechnol 23(4):432–433
Yang Y, Zhang W, Bayrer JR, Weiss MA (2008) Doublesex and the regulation of sexual dimorphism in Drosophila melanogaster: Structure, function, and mutagenesis of a female-specific domain. J Biol Chem 283:7280–7292
Acknowledgments
This research was supported by the research contract no. 12620/RO of the International Atomic Energy Agency, Vienna, Austria, granted to S. Thanaphum. This study is also a part of a master’s degree for R. Permpoon under the supervision of S. Thanaphum at Mahidol University, Thailand. Thanks to the editorial reviewers for their assistance with this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Permpoon, R., Aketarawong, N. & Thanaphum, S. Isolation and characterization of Doublesex homologues in the Bactrocera species: B. dorsalis (Hendel) and B. correcta (Bezzi) and their putative promoter regulatory regions. Genetica 139, 113–127 (2011). https://doi.org/10.1007/s10709-010-9508-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-010-9508-2