Abstract
The neuropeptide B/W signaling system is composed of neuropeptide B (NPB), neuropeptide W (NPW), and two cognate receptors, NPBWR1 and NPBWR2, which are involved in diverse physiological processes, including the central regulation of neuroendocrine axes in vertebrates. The components of this signaling system are not well conserved during vertebrate evolution, implicating its functional diversity. The present study characterized the ricefield eel neuropeptide B/W system, generated a specific antiserum against the neuropeptide B/W receptor, and examined the potential roles of the system in the regulation of adenohypophysial functions. The ricefield eel genome contains npba, npbb, and npbwr2b but lacks the npw, npbwr1, and npbwr2a genes. The loss of npw and npbwr1 probably occurred at the base of ray-finned fish radiation and that of npbwr2a species specifically in ray-finned fish. Npba and npbb genes are produced through whole-genome duplication (WGD) in ray-finned fish. The ricefield eel npba was expressed in the brain and some peripheral tissues, while npbb was predominantly expressed in the brain. The ricefield eel npbwr2b was also expressed in the brain and in some peripheral tissues, such as the pituitary, gonad, heart, and eye. Immunoreactive Npbwr2b was shown to be localized to Lh and Fsh cells but not to Gh or Prl cells in the pituitary of ricefield eels. Npba upregulated the expression of fshb and cga but not lhb mRNA in pituitary fragments of ricefield eels cultured in vitro. The results of the present study suggest that the NPB system of ricefield eels may be involved in the neuroendocrine regulation of reproduction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The neuropeptide B/W signaling system is composed of neuropeptide B (NPB), neuropeptide W (NPW), and their cognate receptors, neuropeptide B/W receptor 1 (NPBWR1) and receptor 2 (NPBWR2). NPB and NPW are structurally similar and synthesized as precursor proteins. The prepro-NPB sequence has two pairs of Arg-Arg motifs at positions 24–25 and 30–31, which could be processed to the NPB mature peptide containing either 23 amino acid residues (NPB23) or 29 amino acid residues (NPB29) after excision of the signal peptide and cleavage at the Arg-Arg motif (Singh and Davenport 2006). However, there are some structural variations in vertebrate NPB peptides. NPB23 is not present in rats and mice due to the lack of the Arg-Arg motif at positions 24–25 in prepro-NPB (Chottova Dvorakova 2018). In addition, chicken NPB lacks one amino acid and forms a special neuropeptide consisting of 28 amino acids (Bu et al. 2016). The prepro-NPW sequence also contains two pairs of Arg-Arg motifs at positions 24–25 and 31–32, which could be processed to the NPW mature peptide containing either 23 amino acid residues (NPW23) or 30 amino acid residues (NPW30) after excision of the signal peptide and cleavage at the Arg-Arg motif. The name NPW is derived from the conserved tryptophan residue (single-letter code W) at the N-terminus and C-terminus of the long-chain form of the mature peptide.
NPBWR1 and NPBWR2, originally named GPR7 and GPR8, are G protein–coupled receptors that show high affinities to NPB and NPW (Chottova Dvorakova 2018). NPBWRs are considered to be coupled to Gi proteins because their activation reduces intracellular forskolin-stimulated cAMP accumulation (Shimomura et al. 2002). Npbwr2 was not found in rodents (Lee et al. 1999), which suggests that the NPB/NPW signaling system has been highly diversified during vertebrate evolution (Chottova Dvorakova 2018). In addition, Npw and Npbwr1 were not found in zebrafish (Bu et al. 2016), which indicates that more dramatic changes have occurred in the composition of the fish NPB/NPW signaling system.
NPB, NPW, and NPBWR are widely distributed in the central nervous system and peripheral tissues in mammals (Chottova Dvorakova 2018; Dun et al. 2005; Fujii et al. 2002; Jackson et al. 2006), chickens (Bu et al. 2016), and fish (Hiraki et al. 2014; Hollander-Cohen et al. 2021; Yang et al. 2014). It has been shown that the NPB/NPW signaling system may regulate food intake and energy balance (Chottova Dvorakova 2018; Ishii et al. 2003; Kelly et al. 2005; Lee et al. 1999; Mondal et al. 2003; Shimomura et al. 2002; Singh et al. 2004; Tanaka et al. 2003), pain signaling (Kelly et al. 2005; Tanaka et al. 2003; Yamamoto et al. 2005; Zaratin et al. 2005), and emotion and behavior (Hiraki-Kajiyama et al. 2019; Kitamura et al. 2006; Lee et al. 1999; Nagata-Kuroiwa et al. 2011; Tanaka et al. 2003). In addition, it may also be involved in endocrine regulation (Bu et al. 2016; Hochol et al. 2007; Price et al. 2009; Samson et al. 2004; Yang et al. 2018a). NPB/NPW signals have been demonstrated to be associated with the regulation of prolactin and growth hormone in rats (Samson et al. 2004), chickens (Bu et al. 2016), and tilapia (Yang et al. 2014) and the regulation of ACTH in rats (Hochol et al. 2007) and chickens (Liu et al. 2022). Interestingly, NPBWR was shown to be enriched in gonadotropes of tilapia; however, whether NPB/NPW directly regulates Lh and/or Fsh remains to be elucidated (Hollander-Cohen et al. 2021).
The ricefield eel (Monopterus albus) is an economically important freshwater fish cultured in China. However, ricefield eel fries for aquaculture are mainly obtained through catching from wild environments due to limited success in large-scale artificial breeding (Feng et al. 2017). Pituitary gonadotropins are key hormones in reproductive regulation in teleosts, and elucidation of the mechanisms underlying the synthesis and release of gonadotropins may aid in the development of techniques for artificial breeding and promote the sustainable aquaculture of ricefield eels. The presence of Npbwr in the gonadotropes of tilapia (Hollander-Cohen et al. 2021) prompted us to examine the potential involvement of the NPB/NPW signaling system in the regulation of Lh and Fsh in ricefield eels. The present study identified three members of the neuropeptide B/W system in the ricefield eel, including NPBa, NPBb, and NPBWR2b, and generated a specific antiserum against Npbwr2b. Fluorescent immunostaining showed that immunoreactive Npbwr2b is present in some Lh and Fsh cells. Npb treatment stimulated the expression of fshb but not lhb in pituitary fragments of ricefield eels cultured in vitro. The present results suggest possible roles for NPB signals in the neuroendocrine regulation of reproduction in ricefield eels.
Materials and methods
Animals and tissues
The adult ricefield eels (body length 30–45 cm and body weight 30–60 g) used for the present study were purchased from a local dealer in Guangzhou, Guangdong, China. Fish were sacrificed by decapitation, after which the pituitary gland and other tissues, including the brain, ovary, testis, muscle, spleen, pancreas, heart, liver, kidney, intestines, blood, eye, and urinary bladder, were dissected out and snap-frozen in liquid nitrogen and stored at − 80 °C for tissue extraction or RNA isolation. The pituitary glands for in vitro culture were directly put in an ice-cold DMEM medium (Gibco, MA, USA). The pituitary glands for histology and immunohistochemistry were fixed in Bouin’s solution for 24 h and stored in 70% ethanol until processing. All procedures and investigations were reviewed and approved by the Center for Laboratory Animals of Sun Yat-sen University and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals.
Chemicals, peptides, and antibodies
DMEM was purchased from Gibco. All the other chemicals were obtained from Sigma Chemical Co. (St. Louis, MO, USA). Ricefield eel Npba23 (WYKQSTGPSYYSVGRASGLLSGI), Npba29 (WYKQSTGPSYYSVGRASGLLSGIRRSPYV), Npbb23 (WYKQVDGPSYYSVGRASGLLSGI), and Npbb29 (WYKQVDGPSYYSVGRASGLLSGIRRSPYV) were synthesized by ChinaPeptides Co., Ltd. (Shanghai, China). The antigen peptide of Npbwr2b (ATVRSKRMPYRTYRAAKI) was synthesized by GL Biochem Ltd. (Shanghai, China). The purity of the synthesized peptides was more than 95% (analyzed by HPLC), and their structures were verified by mass spectrometry. Mouse anti-Actb (actin, beta) monoclonal antibody was purchased from ProteinTech Group, Inc. (IL, USA), and polyclonal antiserum against recombinant Lhb, Fshb, Gh, or Prl of ricefield eel was prepared previously in our laboratory (Yang et al. 2018b, 2021; Wu et al. 2012; Chen et al. 2015).
Sequence analysis
The transmembrane domains of Npbwr were predicted with the online software TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). The signal peptide regions of Npb/Npw precursor protein sequences were predicted with the online software SignalP 5.0 (https://services.healthtech.dtu.dk/service.php? Signalp-5.0). Multiple sequence alignment was performed by Clustalx1.83. The phylogenetic tree was constructed via MEGA 7.0. The amino acid sequences of Npba, Npbb, Npbwr1, and Npbwr2 of representative species were downloaded from NCBI. Information on all genes in the synteny analysis was obtained through the Gene Data Viewer tool in the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/?term =), and the collinear genes around the npba, npbb, npbwr1, and npbwr2 genes were plotted with CorelDRAW 2018 software.
Total RNA extraction
Total RNA was extracted from frozen tissues or cultured pituitary fragments using TRIzol reagent (Invitrogen, MA, USA). The RNA samples were quantified based on the absorbance at 260 nm in a UV/visible spectrophotometer (Amersham Biosciences, Buckinghamshire, England). The A260/280 ratios for all RNA samples were between 1.9 and 2.0. The integrity of RNA was checked with agarose gel electrophoresis and further verified by the amplification of actb (AY647143.1).
RT‒PCR analysis of npba, npbb, and npbwr2b mRNA expression in tissues
One microgram of total RNA from tissues was treated with RNase-free DNase I (Thermo Scientific, MA, USA) to remove genomic DNA contamination and then reverse transcribed using random hexamer primers with the Revert Aid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions. Then, 1 µL of cDNA was used as the template for PCR amplification to detect mRNA expression of each target gene in tissues using the TGRADIENT thermocycler (Biometra GmbH). PCR was performed in a 25-μL final volume containing 2.5 μL 10 X Taq buffer, 2.0 mM MgCl2, 0.2 mM dNTP, 0.4 μM of each primer, and 1.25 units of Taq DNA polymerase (Fermentas, Vilnius, Lithuania). The primers were npba-SQ-F and npba-SQ-R for npba, npbb-SQ-F and npbb-SQ-R for npbb, enpbwr2b-SQ-F and enpbwr2b-SQ-R for enpbwr2b, and b-actin-F2 and b-actin-R2 for actb. The sequences of the primers are listed in Table 1. The PCR mixture was heated at 94 ℃ for 0.5 min, followed by 28 cycles of amplification for actb and 35 cycles for npba, npbb, and npbwr2b. The cycling conditions were 94 °C for 0.5 min, 56 °C for 0.5 min, and 72 °C for 0.5 min, with a final extension at 72 °C for 5 min. The PCR products were separated on a 1.5% agarose gel and captured with the G:BOX F3 Gel Documentation System (Syngene, Cambridge, England).
Generation of the polyclonal antiserum against ricefield eel Npbwr2b
Rabbit antiserum against ricefield eel Npbwr2b (XP_020470699.1) was generated by immunizing rabbits with a synthetic antigen peptide of Npbwr2b (ATVRSKRMPYRTYRAAKI; aa149-166) conjugated to keyhole limpet hemocyanin (KLH) as previously reported (Hu et al. 2002). To examine the specificity of the rabbit anti-Npbwr2b antiserum generated in the present study, the cDNA sequence encoding the C-terminal peptide of ricefield eel Npbwr2b (aa 146–337; designated Npbwr2b-C), which contains the synthetic antigen sequence, was PCR amplified with the primer set enpbwr2-C-F/enpbwr2-C-R and cloned into the prokaryotic expression vector pET-15b. The recombinant Npbwr2b-C protein was expressed in E. coli BL21(DE3) after induction by IPTG. In addition, the cDNA sequence encoding the mature protein of Npbwr2b was PCR amplified with the primer set enpbwr2-ORF-F/enpbwr2-ORF-R and cloned into the eukaryotic expression vector pcDNA3.0. The eukaryotic expression vector was transfected into COS-7 cells to obtain the recombinant mature protein of Npbwr2b. The primers used in the construction of the expression vectors are listed in Table 1. The above recombinant proteins were used in Western blot and immunohistochemical analyses to confirm the specificity of the antiserum.
Western blot analysis
The recombinant proteins or tissue homogenates were separated by electrophoresis on 12% SDS‒PAGE gels and transferred to methanol-activated polyvinylidene fluoride membranes (Merck Millipore, MA, USA) by electroblotting. The membranes were then blocked with 5% nonfat milk powder in 0.01 M PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) at room temperature for 3 h. The blocked membranes were then incubated with rabbit anti-Npbwr2b antiserum (1:1000 dilution) or mouse anti-Actb monoclonal antibody (1:2000, 60,008–1-Ig; ProteinTech Group, Inc., IL, USA) in blocking solution (5% nonfat milk in 0.01 M PBS) at 4 ℃ for 12 h, washed three times with 0.01 M PBS containing 0.05% Tween 20 (PBST) for 10 min, and developed using a 1:1000 dilution of horseradish peroxidase (HRP)–conjugated goat anti-rabbit IgG (H + L) (Beyotime, Shanghai, China) or goat anti-mouse IgG (H + L) (Beyotime) for 1 h at room temperature. After three 10-min final washes with PBST, the membranes were exposed to a chemiluminescence substrate (Super ECL Detection Reagent Kit, 36208ES60, Yeasen, Shanghai, China) according to the manufacturer’s instructions. To confirm the specificity of the Npbwr2b antisera, the control membrane was incubated with the primary antiserum preadsorbed with an excess of recombinant Npbwr2b-C.
Immunohistochemistry
Serial Sections (4–5 µm thick) were cut from Bouin’s-fixed, paraffin-embedded ricefield eel pituitary glands and mounted on glass slides coated with poly-L-lysine. The pituitary sections were deparaffinized, hydrated, and incubated with 3% hydrogen peroxide solution to quench endogenous peroxidase activity, followed by antigen retrieval in 10 mM citrate buffer (pH 6.0) at 95 °C for 15 min and blocking in 0.01 M PBS containing 10% normal goat serum for 30 min at room temperature. For single immunohistochemistry, the blocked sections were incubated with the primary rabbit anti-Npbwr2b antiserum (1:500 dilution) at 4 °C for 12 h. After rinsing with PBS for 5 min three times, the sections were exposed to the secondary antibody (HRP-conjugated goat anti-rabbit IgG (H + L); 1:1000 dilution; Beyotime). After rinsing with PBS, the sections were developed with 3,3’-diaminobenzidine (DAB) solution, mounted, and digitally photographed with a microscope (Eclipse Ni-U, Nikon, Japan). To further confirm the specificity of the immunostaining, control sections were incubated with the primary antiserum (in its working solution) preadsorbed with 200 μg of the recombinant Npbwr2b-C protein.
For double-label fluorescent immunohistochemistry, the blocked sections were incubated in a primary antiserum mixture of rabbit anti-Npbwr2b antiserum (1:500 dilution) with mouse anti-Fshb antiserum (1:1000 dilution), mouse anti-Lhb antiserum (1:500 dilution), mouse anti-Gh antiserum (1:1000 dilution), or mouse anti-Prl antiserum (1:500 dilution) either at 4℃ for 12 h or at 37 ℃ for 2 h. After rinsing with PBS for 5 min three times, the sections were then incubated with secondary antibodies at room temperature for 1 h, using 1:1000 diluted Alexa Fluor 488 labeled Goat Anti-rabbit IgG (A0423; Beyotime) for Npbwr2b, or 1:1000 diluted Cy3 labeled goat anti-mouse IgG (A0521; Beyotime) for Fshb, Lhb, Gh, and Prl. After rinsing with PBS for 5 min three times, 2-[4-amidinopheny]-6-indolecarbamidine (DAPI; 10 µg/mL) was used to stain the nuclei blue. The sections were rinsed with PBS for 5 min three times again and then coverslipped using antifade mounting medium (P0126; Beyotime). Fluorescent signals were photographed with a fluorescence microscope (Eclipse Ti2-E, Nikon, Japan).
In vitro treatment of pituitary fragments of ricefield eel with Npba peptides
The pituitary glands of approximately 200 female, intersexual, and male ricefield eels (body length 40–50 cm and body weight 60–120 g; mixed sexes) in the nonreproductive season were removed and chopped into pieces of approximately 1 mm3 in D-Hanks solution. Pituitary fragments roughly equal to 3 pituitary glands were placed in each well of a 24-well tissue culture plate (Nunc, Denmark) with 0.5 ml of L-15 medium (L4386; Sigma) containing 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco) and then cultured at 28 °C in a humidified incubator (SPX-250BSH-II; CIMO, Shanghai, China). After preculture for 24 h, the medium was replaced, and the pituitary fragments were treated with ricefield eel Npb peptides for the duration indicated in the respective figure legends. Four replicates were performed for each treatment. At the end of treatment, pituitary fragments were collected, and total RNA was extracted for subsequent real-time quantitative PCR analysis.
Quantitative real-time PCR analysis
Total RNA (approximately 500 ng) isolated from cultured pituitary fragments was first treated with RNase-free DNase I (Thermo Scientific) and reverse transcribed with random hexamer primers by using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Then, 1 μL of cDNA template was used for real-time quantitative PCR analysis of cga (Gene ID: 109,955,066), fshb (Gene ID: 109,965,078), lhb (Gene ID: 109,963,826), gh (Gene ID: 109,972,673), and prl (Gene ID: 109,951,456) mRNA levels. The three housekeeping genes, namely, actb, gapdh, and hprt1, were employed as reference genes for qPCR analysis by following the suggestions of a previous report (Vandesompele et al. 2002), which is intended to minimize misestimating mRNA expression levels through qPCR due to the potential variations in the expression levels of any single reference gene. The geometric mean expression levels of the three reference genes were used to normalize the expression levels of the target genes. The primers were hprt1-QF and hprt1-QR for hprt1, β-actin-QF and β-actin-QR for actb, gapdh-QF and gapdh-QR for gapdh, lhb-QF and lhb-QR for lhb, fshb-QF and fshb-QR for fshb, cga-QF and cga-QR for cga, gh-QF and gh-QR for gh, and prl-QF and prl-QR for prl. The nucleotide sequences of these primers are listed in Table 1. qPCR analysis was performed in a 10 µL reaction volume containing 0.3 µM of each primer, 1 µL of cDNA template, and 5 µL of Bestar® SybrGreen qPCR mastermix (DBI® Bioscience, Germany) using a Roche LightCycler 480 detection system. The cycling conditions were 5 min at 95 °C and 40 cycles of 10 s at 95 °C, 15 s at 58 °C and 20 s at 72 °C. Data were analyzed by LightCycler 480 software. The specificity of qPCR amplification was confirmed by melt-curve analysis, gel electrophoresis, and sequencing of PCR products. All samples were run in duplicate, and minus reverse transcriptase and no template controls were included in each assay.
Quantification of the mRNA expression levels was performed using a standard curve with tenfold serial dilutions of plasmid containing the corresponding DNA fragment, which ranged from 101 to 108 copies, and the PCR efficiencies were between 1.9 and 2.05. The copy numbers of genes were calculated by LightCycler 480 software based on the corresponding standard curves. The mRNA expression levels of fshb, lhb, cga, gh, and prl are presented as the copy number ratios to the geometric means of the reference genes.
Data analysis
All data are expressed as the mean ± SEM. The significance of observed differences between groups was determined by one-way ANOVA followed by Tukey’s multiple comparison test using GraphPad Prism software. The statistical significance was set at p < 0.05.
Results
Sequence analysis of ricefield eel npba, npbb, and npbwr2b
We identified npba (Gene ID: 109,957,844), npbb (Gene ID: 109,972,237), and npbwr2b (Gene ID: 109,968,640) but not npw, npbwr1, or npbwr2a in the M. albus genome (Genome ID: 24,053). The npba, npbb, and npbwr2b genes of M. albus consist of one intron and two exons, which encode putative precursor proteins of 130, 126, and 337 amino acids, respectively. The amino acid sequence identity was 72.3% for the Npba and Npbb precursors.
The signal peptide regions of ricefield eel Npba and Npbb precursors were predicted with signalP-5.0 online software, which is not conserved among species (Online Resource 1). The mature peptides of ricefield eel Npba and Npbb start with the amino acid residue W and contain two “RR” motifs at the C-terminal portions, which may produce two potential mature peptides containing either 23 or 29 amino acids after enzymatic processing. The mature peptides of ricefield eel Npba and Npbb shared a high sequence identity, with only two different amino acids at positions 5 and 6 (serine and threonine for Npba and valine and aspartic acid for Npbb; Online Resource 1).
Ricefield eel Npbwr2b is a GPCR that contains seven transmembrane domains. Except for the N-terminal extracellular and C-terminal intracellular regions, the sequences of NPBWRs among different species are relatively conserved (Online Resource 2).
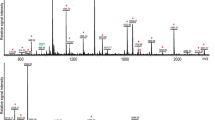
Phylogenetic analysis showed that vertebrate NPB/NPW can be categorized into three major groups, namely, NPB of cartilaginous fishes, ray-finned fishes, lobe-finned fishes, and amphibians; NPB of reptiles, birds, and mammals; and NPW of cartilaginous fishes, lobe-finned fishes, amphibians, birds, and mammals (Fig. 1). Ray-finned fish Npba and Npbb was categorized into two sister groups along the branch of NPB of cartilaginous fishes, ray-finned fishes, lobe-finned fishes, and amphibians. Ricefield eel Npba and Npbb were closely related to the counterparts of medaka and tilapia, respectively. The Npb-L sequence of elephant shark was clustered with NPW in the phylogenetic tree.
Phylogenetic analysis of NPBs/NPWs in vertebrates. The phylogenetic tree was constructed with the neighbor-joining method in the MEGA7 software. Lamprey Npb was used as an outgroup to root the tree. The bootstrap values (%, 1000 replicates) were given at each branch point. Vertebrate NPBs/NPWs could be categorized into three major groups. The ricefield eel Npba and Npbb were marked with black dots, which were clustered with teleost fish Npba and Npbb, respectively
Vertebrate NPBWR can be categorized into four groups, namely, NPBWR1 of mammals, NPBWR2 of mammals, NPBWR2 of cartilaginous fishes, lobe-finned fishes, teleosts, amphibians, reptiles, and birds, and NPBWR1 of cartilaginous fishes, lobe-finned fishes, amphibians, reptiles, and birds (Fig. 2). Ricefield eel Npbwr2b is closely related to that of sea bass.
Phylogenetic analysis of NPBWRs in vertebrates. The phylogenetic tree was constructed with the neighbor-joining method in the MEGA7 software. Lamprey Npbwrs were used as outgroups to root the tree, which was collapsed for simplification. The bootstrap values (%, 1000 replicates) were given at each branch point. Branches corresponding to partitions reproduced in less than 70% of bootstrap replicates are collapsed. Vertebrate NPBWRs were categorized into four groups. The ricefield eel Npbwr2b was marked with a black dot, which was clustered with non-mammalian Npbwr2s, especially closely related to teleost Npbwr2s
Synteny analysis of ricefield eel npba, npbb, and npbwr2b
To ascertain the absence of npw, npbwr1, and npbwr2a in the ricefield eel genome, synteny analysis was performed by searching the neighboring genes around npb/npw or npbwr in the genomes of lamprey, elephant shark, lobe-finned fishes, and ray-finned fishes. The conserved syntenic regions around npb were identified in elephant shark, lobe-finned fishes, and ray-finned fishes. The npw gene exists in the genome of elephant shark and lobe-finned fishes but is absent in the genome of the ray-finned fishes examined, including zebrafish, striped catfish, striped sea bass, Japanese medaka, and ricefield eel (Fig. 3). Some of the neighboring genes around npw of lobe-finned fishes could also be identified in the genomes of ray-finned fishes, but they are separated into two different chromosomes or contigs (Fig. 3).
Synteny analysis of npb/npw genes in lamprey and fishes. Location of npb/npw genes was identified by the Gene Data Viewer tool in the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/?term =). The diagram was drawn with the CorelDRAW 2018 software. Dotted lines indicate the syntenic genes; dashed lines denote genes of interest. Only one npb gene exists in lampreys, elephant shark, lobe-finned fishes (coelacanth and west African lungfish), and some ray-finned fish (zebrafish and striped sea bass), but two npb genes in some other ray-finned fish (striped catfish, Japanese medaka, and ricefield eel). One npw gene exists in elephant shark and lobe-finned fish (coelacanth and west African lungfish), but none in ray-finned fish. All the genomic data used were downloaded from Genomicus (http://www.genomicus.biologie) and Esemble Genome Browser (http://www.ensembl.org). Sea lamprey, Petromyzon marinus; Elephant shark, Callorhinchus milii; Coelacanth, Latimeria chalumnae; West African lungfish, Protopterus annectens; Zebrafish, Danio rerio; Striped catfish, Pangasianodon hypophthalmus; Striped sea bass, Morone saxatilis; Medaka, Oryzias latipes; Ricefield eel, Monopterus albus. Chr: chromosome
The npbb gene exists in some ray-finned fishes such as striped catfish, Japanese medaka, and ricefield eel but is absent in some other ray-finned fishes such as zebrafish and striped sea bass (Fig. 3). The npba and npbb genes of striped catfish, Japanese medaka, and ricefield eel are present on two different chromosomes or contigs, and both showed conserved synteny with npb of lobe-finned fishes and the elephant shark (Fig. 3). Unexpectedly, the elephant shark npb-l showed conserved synteny with npw of lobe-finned fishes (Fig. 3).
The npbwr1 gene exists in the elephant shark and lobe-finned fishes but is absent in the examined ray-finned fishes (Fig. 4). However, some neighboring genes around the npbwr1 genes of the elephant shark and lobe-finned fishes are also present in the genomes of the ray-finned fishes. One npbwr2 homolog gene was identified in the genomes of elephant shark, lobe-finned fishes, and some ray-finned fishes, such as striped sea bass, Japanese medaka, and ricefield eel. In contrast, two npbwr2 homolog genes, namely, npbwr2a and npbwr2b, are present in some other ray-finned fishes, such as zebrafish and striped catfish. These npbwr2 homologs showed conserved synteny, and some of the syntenic genes around npbwr2a could also be identified in striped sea bass, Japanese medaka, and ricefield eel, although their npbwr2a genes are absent (Fig. 4). The npbwr2a and npbwr2b genes of zebrafish and striped catfish are located on the same chromosome. The syntenic regions of npbwr2 homologs in striped sea bass and Japanese medaka are also located on the same chromosome or contig. In the ricefield eel, the syntenic regions of npbwr2 homologs seemed to be on different contigs, which is likely an artifact due to low assembly quality in these regions. For the two npbwr2 genes, it appears that npbwr2a was under less evolutionary pressure with relatively high sequence variations. However, npbwr2b was under higher evolutionary pressure with highly conserved sequences.
Synteny analysis of npbwr genes in lamprey and fishes. Location of npbwr genes was identified by the Gene Data Viewer tool in the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/?term =). The diagram was drawn with the CorelDRAW 2018 software. Dotted lines indicate the syntenic genes; dashed lines denote genes of interest. There were three npbwr1 genes in lampreys, one npbwr1 gene in elephant shark and lobe-finned fishes (coelacanth and west African lungfish), but none in ray-finned fish. Only one npbwr2 gene exists in elephant shark, lobe-finned fishes, and some ray-finned fishes (striped sea bass, Japanese medaka, and ricefield eel), but two exist in other ray-finned fishes (zebrafish and striped catfish) which are located on the same chromosome. Sea lamprey, Petromyzon marinus; Elephant shark, Callorhinchus milii; Coelacanth, Latimeria chalumnae; West African lungfish, Protopterus annectens; Zebrafish, Danio rerio; Striped catfish, Pangasianodon hypophthalmus; Striped sea bass, Morone saxatilis; Medaka, Oryzias latipes; Ricefield eel, Monopterus albus. Chr: chromosome
mRNA expression of npba, npbb, and npbwr2b in ricefield eel tissues
The expression of npba, npbb, and npbwr2b in tissues of female and male ricefield eels was examined with RT‒PCR analysis. The results showed that npba was expressed in the brain as well as some peripheral tissues of female (Fig. 5) and male (Fig. 6) fish. In females, npba was expressed at relatively higher levels in the hypothalamus and medulla, and at relatively lower levels in the telencephalon, optic tectum-thalamus, cerebellum, spleen, pancreas, blood cell, and urinary bladder but barely or not detectable in the olfactory bulb, pituitary, gonad, muscle, heart, liver, kidney, intestine, and eye. In males, ricefield eel npba was expressed at relatively higher levels in the olfactory bulb, hypothalamus, optic tectum-thalamus, cerebellum, and medulla, and at relatively lower levels in the telencephalon, blood cell, and eyes but barely or not detectable in the pituitary, gonad, muscle, spleen, pancreas, liver, kidney, heart, intestine, and urinary bladder.
RT-PCR analysis of npba, npbb, and npbwr2b mRNA expression in tissues of female ricefield eels. Ob, olfactory bulb; Te, telencephalon; Hy, hypothalamus; Ot, optic tectum-thalamus; Ce, cerebellum; Mo, medulla oblongata; Go, Gonad; Mu, muscle; Sp, spleen; Pa, pancreas; He, heart; Li, liver; Ki, kidney; In, intestines; Bl, blood cells; Ey, eyes; Ub, urinary bladder; RT-, RT minus (no addition of reverse transcriptase); NC, negative control (water used as template)
RT-PCR analysis of npba, npbb, and npbwr2 mRNA expression in tissues of male ricefield eels. Ob, olfactory bulb; Te, telencephalon; Hy, hypothalamus; Ot, optic tectum-thalamus; Ce, cerebellum; Mo, medulla oblongata; Go, Gonad; Mu, muscle; Sp, spleen; Pa, pancreas; He, heart; Li, liver; Ki, kidney; In, intestines; Bl, blood cells; Ey, eyes; Ub, urinary bladder; RT-, RT minus (no addition of reverse transcriptase); NC, negative control (water used as template)
npbb mRNA was detected predominantly in the central nervous system of female (Fig. 5) and male (Fig. 6) fish. In females, npbb was expressed at relatively higher levels in the hypothalamus, cerebellum, and medulla, and at a lower level in the telencephalon, optic tectum-thalamus, and eyes but was not detectable in other tissues examined. In males, npbb was expressed predominantly in the hypothalamus, optic tectum-thalamus, and medulla but barely or not detectable in other tissues examined.
npbwr2b seemed to be expressed more widely in tissues of female fish (Fig. 5) than in those of male fish (Fig. 6). In females, npbwr2b was detected at relatively higher levels in the telencephalon, hypothalamus, optic tectum-thalamus, cerebellum, medulla, and pituitary and at relatively lower levels in the gonad, spleen, pancreas, heart, kidney, intestine, and eye but barely or not detectable in the olfactory bulb, muscle, liver, blood cell, and urinary bladder. In males, npbwr2b was detected at relatively higher levels in the telencephalon, hypothalamus, optic tectum-thalamus, medulla, and pituitary, and at relatively lower levels in the olfactory bulb, cerebellum, gonad, heart, and eye but barely or not detectable in other tissues examined.
Generation of the rabbit anti-ricefield eel Npbwr2b antiserum
Rabbit anti-Npbwr2b antiserum was generated against a synthetic peptide corresponding to the second intracellular region of ricefield eel Npbwr2b (aa149-166; ATVRSKRMPYRTYRAAKI). Western blot analysis showed that the rabbit anti-Npbwr2b antiserum could recognize the recombinant ricefield eel Npbwr2b-C peptide expressed in E. coli (Fig. 7A) and the full-length ricefield eel Npbwr2b protein expressed in COS-7 cells (Fig. 7B). The signals disappeared when the antiserum was preadsorbed by the recombinant ricefield eel Npbwr2b-C peptide (Fig. 7C and D). These results suggest that the rabbit anti-Npbwr2b antiserum is of high specificity.
The specificity of the rabbit anti-Npbwr2b antiserum as determined by Western blot analysis. The recombinant Npbwr2b-C in bacterial extracts (10 μg; A and C) or the recombinant full-length Npbwr2b in COS-7 cell extracts (80 µg; B and D) were separated on 12% SDS-PAGE gels, transferred to polyvinylidene fluoride membranes, and immunoreacted with the rabbit anti-Npbwr2b antiserum (1:1000 dilution; A and B) or the anti-Npbwr2b antiserum (1:1000 dilution) preadsorbed with 50 μg /mL of Npbwr2b-C (C and D). The secondary antibody was 1:1000 diluted horseradish peroxidase (HRP)–conjugated goat anti-rabbit IgG (H + L) (Beyotime, Shanghai, China, A0208), and the blots were visualized using the super ECL Detection Reagent Kit (Yeasen; 36208ES20). BL21(DE3): bacterial protein extracts of BL21(DE3). pET-15b: bacterial protein extracts of BL21(DE3) transformed with the empty pET-15b vector. Npbwr2b-C: bacterial protein extracts of BL21(DE3) transformed with the expression construct pET-15b-Npbwr2b-C. COS-7: protein extracts of COS-7 cells. pcDNA3: protein extracts of COS-7 cells transfected with the empty pcDNA3 vector. Npbwr2b: protein extracts of COS-7 cells transfected with the expression construct pcDNA3-Npbwr2b
Western blot analysis of tissue homogenates revealed an immunoreactive band of approximately 38 kD in the brain, ovary, kidney, and pancreas but not in the liver and muscle, which disappeared when the antiserum was preadsorbed by the recombinant ricefield eel Npbwr2b-C peptide (Fig. 8). These results further suggest high specificity of the antiserum generated in the present study.
Western blot analysis of immunoreactive Npbwr2b in tissues of female Monopterus albus. The tissue homogenates (10 μg) prepared with the RIPA lysis buffer (Beyotime, Shanghai, China) from the brain (Br), ovary (Ov), kidney (Ki), liver (Li), pancreas (Pa), and muscle (Mu) were separated on 12% SDS-PAGE gels, transferred to polyvinylidenefluoride membranes, and immunoreacted with the rabbit anti-Npbwr2b antiserum (1:1000 dilution; A), or the anti-Npbwr2b antiserum (1:1000 dilution) preadsorbed with 50 μg/mL of Npbwr2b-C (B), or mouse anti-Actb monoclonal antibody (1:20,000; catalog number: 60008–1-Ig; ProteinTech Group, Inc.; C). The secondary antibody was 1:1000 diluted horseradish peroxidase (HRP)–conjugated goat anti-rabbit IgG (H + L) (Beyotime, Shanghai, China, A0208) or horseradish peroxidase (HRP)–conjugated goat anti-mouse IgG (H + L) (catalog number 115–035-003; Jackson ImmunoResearch Laboratories). The blots were visualized using the super ECL Detection Reagent Kit (YEASEN, 36208ES20)
Cellular localization of immunoreactive Npbwr2b in the pituitary of ricefield eels
Immunohistochemical analysis of the pituitary gland of ricefield eels showed that Npbwr2b immunostaining was mainly localized to the periphery of the pituitary gland, namely, the adenohypophysis (Fig. 9A and A1). The immunopositive signals were abolished when the antiserum was preadsorbed by the recombinant Npbwr2b-C peptide (Fig. 9B and B1), suggesting the specificity of the immunopositive Npbwr2b signals in the pituitary. The cellular localization of Npbwr2b in the pituitary gland was further examined with double fluorescence immunohistochemistry. Immunoreactive signals of Npbwr2b were shown to be colocalized with either Fsh (Fig. 10A and A1) or Lh cells (Fig. 10B and B1). No Npbwr2b immunostaining was detected in either Gh cells (Fig. 10C and C1) or Prl cells (Fig. 10D and D1).
Cellular localization of immunoreactive Npbwr2b in the pituitary of female ricefield eels. The rabbit anti-Npbwr2b antiserum (1:1000, A), or the preabsorbed rabbit anti-Npbwr2b antiserum (1:1000, B) by the excessive recombinant Npbwr2b-C peptide (50 μg/mL) was used as the primary antiserum. The secondary antibody was 1:1000 diluted horseradish peroxidase (HRP)–conjugated goat anti-rabbit IgG (H + L) (Beyotime, Shanghai, China; A0208), and the sections were visualized using the DAB kit (MXB, DAB-0031). A1 and B1 are the higher magnification of the boxed areas in A and B, respectively. The scale bar represents 20 μm
Co-localization of immunoreactive Npbwr2b (green) with Fshb, Lhb, Gh, or Prl (red) in the pituitary of female ricefield eels. The rabbit antiserum against Npbwr2b (1:500 dilution) and mouse antiserum against Fshb (1:1000 dilution; A), Lhb (1:500 dilution; B), Gh (1:1000 dilution; C), or Prl (1:500 dilution; D) were used as primary antisera. The secondary antibodies were 1:1000 diluted Alexa Fluor 488 labeled goat anti-rabbit IgG (Beyotime, Shanghai, China, A0423) for Npbwr2b and 1:1000 diluted Cy3 labeled goat anti-mouse IgG (Beyotime, Shanghai, China, A0521) for Fshb, Lhb, Gh, and Prl. DAPI was used to stain the nuclei blue. The images were observed and captured with a fluorescence microscope (Eclipse Ti2-E, Nikon, Japan). A1, B1, C1, and D1 are higher magnification of the boxed areas in A, B, C, and D, respectively. The overlapping of the red with the green color generated a yellow color. The scale bar is 20 μm
The effects of Npb on the expression of lhb, fshb, and cga in pituitary fragments of ricefield eels cultured in vitro
As Npbwr2a was shown to be colocalized with Fsh cells and Lh cells in the pituitary of ricefield eels, the potential regulatory roles of NPB signals in Fsh and Lh cells were further examined. In pituitary fragments cultured in vitro, the results of time-course experiments showed that the expression of cga mRNA was significantly increased after 1 h of culture with Npba23 and Npba29 (100 nM; Fig. 11A). The expression of fshb mRNA was significantly increased after 1 h of culture with Npba23 and after 0.5 to 2 h of culture with Npba29 (Fig. 11B). The expression of gh mRNA was significantly increased at 12 h after the addition of Npba23 and at 4 and 12 h after the addition of Npba29 (Fig. 11D). The expression of either lhb or prl was not altered after 0.5 to 12 h of culture with Npba23 or 29 (Fig. 11C and D).
Time-course effects of Npba23 and Npba29 (100 nM) on the expression of cga, fshb, lhb, gh, and prl mRNA in pituitary fragments of mixed sexes of ricefield eels cultured in vitro. The pituitary fragments were collected after treatment with Npba23 or Npba29, and processed for total RNA extraction and qPCR analysis. Data were expressed as fold change relative to the corresponding vehicle control. Bars represent means ± SEM (n = 4). *p < 0.05 vs the vehicle control
The results of dose-dependent studies showed that the expression of cga was increased dose-dependently after 1 h of culture with increasing concentrations of Npba23 and Npba29 (1 to 1000 nM), with a significant difference observed at 1000 nM Npba29 when compared to the control (Fig. 12A). The expression of fshb was also increased dose-dependently after 1 h of culture with increasing concentrations of Npba23 and Npba29, with significant differences observed at 1000 nM for Npba23 and at 100 nM for Npba29 when compared to the control (Fig. 12B). The expression of either lhb or prl was not altered by 1 h culture with increasing concentrations of Npba23 or Npba29 (Fig. 12C and D). Npbb seemed not to alter the expression of cga, fshb, lhb, or prl mRNA (Online Resource 3).
Dose-dependent effects of Npba23 and Npba29 on the expression of cga, fshb, lhb, and prl mRNA in pituitary fragments of mixed sexes of ricefield eels cultured in vitro. The pituitary fragments were collected 1 h after culture with Npba23 or Npba29, and processed for total RNA extraction and qPCR analysis. Data were expressed as fold change relative to the corresponding vehicle control. Bars represent means ± SEM (n = 4). *p < 0.05 vs the vehicle control
Discussion
Members of the neuropeptide B/W signaling system in vertebrates include NPB, NPW, NPBWR1, and NPBWR2, which exhibit lineage-specific differences due to gene duplication and/or loss during evolution. To date, most studies concerning the neuropeptide B/W signaling system have been performed in mammals and chickens. In the present study, the neuropeptide B/W signaling system was characterized in the ricefield eel M. albus, a teleost fish in Synbranchiformes. The ricefield eel genome contains npba, npbb, and npbwr2a but lacks npw and npbwr1. Ricefield eel NPBa, NPBb, and NPBWR2a are closely related to their counterparts in Percomorpha. The npw and npbwr1 genes were also shown to be absent by synteny analysis in other ray-finned fish examined but present in lobe-finned fish and tetrapod vertebrates, suggesting that the loss of npw and npbwr1 genes occurred at the base of ray-finned fish radiation. However, the significance of these evolutionary changes remains to be elucidated. It is of interest to note that although lacking the C-terminal tryptophan residue of vertebrate NPW molecules, the elephant shark Npb-l was phylogenetically clustered with NPW. Moreover, the elephant shark nbp-l gene also showed conserved synteny with the lobe-finned fish npw gene and the putative npw chromosomal region of ray-finned fishes. These lines of evidence suggest that NPW possibly evolved from NPB through an intermediary characteristic of the elephant shark Npb-l.
In contrast to the elephant shark and lobe-finned fishes, two forms of NPB, namely, NPBa and NPBb, are present in some ray-finned fishes, including striped catfish, Japanese medaka, and ricefield eel. The npba and npbb genes of these fish are located on different chromosomes or contigs, and both showed conserved synteny with npb genes of the elephant shark and lobe-finned fishes. These lines of evidence suggest that two copies of npb genes in ray-finned fishes were probably generated through a unique genomic replication event (3R) in ray-finned fish (Hiraki-Kajiyama et al. 2019; Kasahara 2013; Meyer and Van de Peer 2005; Vandepoele et al. 2004). The absence of npbb in zebrafish and striped sea bass is probably due to species-specific gene loss after duplication of npb and suggests that npbb is under less selection pressure than npba during the evolution of ray-finned fishes.
Only one form of npbwr2, namely, npbwr2b, exists in the genomes of ricefield eel, striped sea bass, and Japanese medaka. In contrast, two forms of npbwr2, including npbwr2a and npbwr2b, were identified in some ray-finned fishes, such as zebrafish and striped catfish. Interestingly, in zebrafish and striped catfish, npbwr2a and npbwr2b appear to be present on the same chromosome and showed conserved synteny with npbwr2 genes of elephant shark and lobe-finned fish. Moreover, neighboring genes around zebrafish and striped catfish npbwr2b were also identified on the same chromosome or contig as npbwr2a of striped sea bass and Japanese medaka. These lines of evidence suggest that the npbwr2 gene was probably duplicated to produce npbwr2a and npbwr2b through chromosome fragment replication at the base of ray-finned fish radiation. It is likely that the npbwr2a gene was under less selection pressure and lost species specifically in some ray-finned fishes, including the ricefield eel, striped sea bass, and Japanese medaka.
It has been shown that the Npb gene is highly expressed in the central nervous system as well as in some peripheral tissues of mammals (Chottova Dvorakova 2018) and teleosts (Hiraki et al. 2014; Hiraki-Kajiyama et al. 2019; Yang et al. 2014). Similarly, the mRNA expression of ricefield eel npba and npbb was also detected in brain areas including the hypothalamus and in some peripheral tissues of males and females. Interestingly, ricefield eel npba and npbb showed different tissue-specific expression patterns in the central nervous system as well as the peripheral tissues, with the expression of npba but not npbb detected in the olfactory bulb, spleen, pancreas, blood cells, and urinary bladder. In medaka, the coexpression of npba and npbb was detected in the same neurons in the supracommissural/posterior nucleus of the ventral telencephalic area (Vs/Vp) and the magnocellular/gigantocellular portion of the magnocellular preoptic nucleus (PMm/PMg) but not in other brain nuclei (Hiraki-Kajiyama et al. 2019). These results suggest that after duplication, the npba and npbb genes might have experienced neo- and/or subfunctionalization during evolution in ray-finned fishes. In addition, RT‒PCR analysis showed that the expression of ricefield eel npba was obviously detectable in the olfactory bulb of males but not females and in the urinary bladder of females but not males, and the expression of ricefield eel npbb was detected in the cerebellum and eye of females but not males. In medaka, npba and npbb genes were shown to be female-specifically expressed in the telencephalic and preoptic nuclei of the brain, which results from the direct stimulatory action of ovarian estrogens (Hiraki et al. 2014; Hiraki-Kajiyama et al. 2019). If the sex-dimorphic expression of npba and npbb in tissues of ricefield eels is resulted from direct actions of sex steroids remains to be clarified.
The expression of ricefield eel npbwr2b mRNA was detected in all discrete brain regions of both females and males. Similarly, npbwr2 was also found to be widely expressed in the central nervous system of medaka without overt differences between the sexes (Hiraki-Kajiyama et al. 2019). The predominant expression of npb and npbwr2b in the brains of ricefield eels and medaka is suggestive of important roles for the Npb signaling system in the functional regulation of the central nervous system, which is probably related to food intake (Sakurai 2013; Yang et al. 2014) and mating behaviors (Hiraki-Kajiyama et al. 2019). In addition, the expression of npb and npbwr2b was also detected in some peripheral tissues including the urinary bladder and eye, which suggests that the NPB signaling system may also be involved in the regulation of physiological functions of these peripheral tissues of ricefield eels.
As in other vertebrates (Bu et al. 2016; Hiraki-Kajiyama et al. 2019), npbwr2b mRNA was also detected in the pituitary of ricefield eels by RT‒PCR analysis. The cellular localization of Npbwr2b in the pituitary of ricefield eels was further examined with double fluorescent immunohistochemical analysis, which showed that Npbwr2b immunostaining was colocalized with some Lh and Fsh cells but not with Gh cells or Prl cells. In tilapia, the neuropeptide W/neuropeptide B receptor was also shown to be specifically enriched in gonadotropes (Hollander-Cohen et al. 2021). These lines of evidence suggest that the NPB signaling system may be directly involved in the regulation of pituitary gonadotropes but not Gh or Prl cells in ricefield eels. In agreement, Npba was shown to stimulate the expression of fshb and cga. In contrast, the expression of lhb was not altered by Npba treatment in the pituitary fragments of ricefield eels cultured in vitro, which may represent one of the neuroendocrine mechanisms underlying the differential regulation of Lh and Fsh in ricefield eels. The roles of the NPB signaling system in Lh cells remain to be identified. These results suggest that NPBa may be involved in the neuroendocrine regulation of reproduction in ricefield eels possibly through upregulation of fshb and cga expression. To further verify the roles of NPBa in reproduction, it is worthwhile to compare the expression levels of npba gene in the hypothalamus and npbwr2b gene in the pituitary of reproductive immature and mature ricefield eels, and to examine if Npba could regulate GnRH synthesis and release in the hypothalamus.
Consistent with the lack of immunoreactive Npbwr2b in Prl cells in the pituitary of ricefield eels, Npba treatment did not alter the expression of prl in the pituitary fragments cultured in vitro. In tilapia, conversely, intraperitoneal injection of Npb stimulated prl expression in the pituitary (Yang et al. 2014). This discrepancy is probably due to the differences in experimental procedures and/or species-specific effects of Npb. Although Gh cells did not express Npbwr2b in the pituitary of ricefield eels, Npba could stimulate gh expression in the pituitary fragments cultured in vitro. Gh expression and release have been shown to be regulated by Lh through the intrapituitary feedback loop in a teleost fish, grass carp (Wong et al. 2006). Thus, it is likely that some Npba-induced factors from Lh and/or Fsh cells stimulate gh expression in the pituitary fragments of ricefield eels cultured in vitro. The response time for Gh cells in increasing gh expression to Npba challenge was much longer than that for Fsh cells of pituitary fragments cultured in vitro, which is probably due to that it takes time for the Npba-induced factors from Lh and/or Fsh cells to accumulate to a high enough concentration to stimulate gh expression. Contrastingly in tilapia, intraperitoneal injection of Npb inhibited gh expression in the pituitary (Yang et al. 2014). In addition to the pituitary, the neuropeptide B/W receptor was also detected in the brain and some peripheral organs such as the eye, gonad, and heart. The intraperitoneal injection of Npb might elicit systemic effects, which resulted in the inhibition of gh expression observed in tilapia (Yang et al. 2014). In contrast in pituitary fragments cultured in vitro, administration of Npba might only activate the receptor Npbwr2b in the pituitary, which resulted in the increase of gh expression observed in ricefield eels in the present study. In chickens, however, Npb could not activate either Npbwr1 or Npbwr2 in vitro (Bu et al. 2016). In contrast, chicken Npw was shown to inhibit the synthesis and secretion of ACTH (Liu et al. 2022) and the secretion of growth hormone (GH) and prolactin (PRL) via neuropeptide B/W receptor 2 (NPBWR2) (Bu et al. 2016). Thus, the functional roles and mechanisms of the NPB/NPW signaling system in the pituitary of vertebrates may vary in different species.
In conclusion, the NPB/NPW signaling system of ricefield eel is composed of NPBa, NPBb, and NPBWR2b. The present results suggest that npw and npbwr1 genes were lost at the base of ray-finned fish radiation, and npbwr2a originated through chromosomal fragment duplication but was subsequently lost species specifically in ray-finned fishes. The npba, npbb, and npbwr2b genes were expressed in various brain regions including the hypothalamus. Immunoreactive Npbwr2b was colocalized with Lh and Fsh cells but not with Gh or Prl cells. Npba is possibly involved in the regulation of fshb and gh but not lhb or prl. The NPB system in the pituitary of ricefield eel may play roles in the neuroendocrine regulation of reproduction possibly through direct upregulation of fshb and cga expression, and of growth possibly through indirect upregulation of gh expression. If Npba acts in the brain to regulate fshb and cga expression via GnRH and/or GnIH and gh expression via GHRH and/or somatostatin remains to be elucidated. The regulation of pituitary functions by the NPB/NPW signaling system and the underlying mechanisms may be diversified in vertebrates.
Data availability
The data presented in this study is contained within the article and supplementary material.
References
Bu GX, Lin DL, Cui L, Huang L, Lv C, Huang SM, Wan YP, Fang C, Li J, Wang YJ (2016) Characterization of neuropeptide B (NPB), neuropeptide W (NPW), and their receptors in chickens: evidence for NPW being a novel inhibitor of pituitary GH and prolactin secretion. Endocrinology 157(9):3562–3576. https://doi.org/10.1210/en.2016-1141
Chen D, Liu J, Chen W, Shi S, Zhang W, Zhang L (2015) Expression and ontogeny of growth hormone (Gh) in the protogynous hermaphroditic ricefield eel (Monopterus albus). Fish Physiol Biochem 41(6):1515–1525. https://doi.org/10.1007/s10695-015-0104-3
Chottova Dvorakova M (2018) Distribution and function of neuropeptides W/B signaling system. Frontiers in Physiology 9:981. https://doi.org/10.3389/Fphys.2018.00981
Dun SL, Brailoiu GC, Mizuo K, Yang J, Chang JK, Dun NJ (2005) Neuropeptide B immunoreactivity in the central nervous system of the rat. Brain Res 1045(1–2):157–163. https://doi.org/10.1016/j.brainres.2005.03.024
Feng K, Luo H, Li Y, Chen J, Wang Y, Sun Y, Zhu Z, Hu W (2017) High efficient gene targeting in rice field eel Monopterus albus by transcription activator-like effector nucleases. Science Bulletin 62(3):162–164. https://doi.org/10.1016/j.scib.2017.01.018
Fujii R, Yoshida H, Fukusumi S, Habata Y, Hosoya M, Kawamata Y, Yano T, Hinuma S, Kitada C, Asami T, Mori M, Fujisawa Y, Fujino M (2002) Identification of a neuropeptide modified with bromine as an endogenous ligand for GPR7. J Biol Chem 277(37):34010–34016. https://doi.org/10.1074/jbc.M205883200
Hiraki T, Nakasone K, Hosono K, Kawabata Y, Nagahama Y, Okubo K (2014) Neuropeptide B is female-specifically expressed in the telencephalic and preoptic nuclei of the medaka brain. Endocrinology 155(3):1021–1032. https://doi.org/10.1210/en.2013-1806
Hiraki-Kajiyama T, Yamashita J, Yokoyama K, Kikuchi Y, Nakajo M, Miyazoe D, Nishiike Y, Ishikawa K, Hosono K, Kawabata-Sakata Y, Ansai S, Kinoshita M, Nagahama Y, Okubo K (2019) Neuropeptide B mediates female sexual receptivity in medaka fish, acting in a female-specific but reversible manner. Elife 8:e39495. https://doi.org/10.7554/eLife.39495
Hochol A, Tortorella C, Rucinski M, Ziolkowska A, Nussdorfer GG, Malendowicz LK (2007) Effects of neuropeptides B and W on the rat pituitary-adrenocortical axis: in vivo and in vitro studies. Int J Mol Med 19(2):207–211
Hollander-Cohen L, Golan M, Levavi-Sivan B (2021) Differential regulation of gonadotropins as revealed by transcriptomes of distinct LH and FSH cells of fish pituitary. International. J Molecular Sci 22(12):6478. https://doi.org/10.3390/Ijms22126478
Hu YX, Guo JY, Shen L, Chen Y, Zhang ZC, Zhang YL (2002) Get effective polyclonal antisera in one month. Cell Res 12(2):157–160
Ishii M, Fei H, Friedman JM (2003) Targeted disruption of GPR7, the endogenous receptor for neuropeptides B and W, leads to metabolic defects and adult-onset obesity. Proc Natl Acad Sci U S A 100(18):10540–10545. https://doi.org/10.1073/pnas.1334189100
Jackson VR, Lin SH, Wang Z, Nothacker HP, Civelli O (2006) A study of the rat neuropeptide B/neuropeptide W system using in situ techniques. J Comp Neurol 497(3):367–383. https://doi.org/10.1002/cne.20989
Kasahara M (2013) Impact of whole-genome duplication on vertebrate development and evolution. Semin Cell Dev Biol 24(2):81–82 (S1084-9521(13)00011-6)
Kelly MA, Beuckmann CT, Williams SC, Sinton CM, Motoike T, Richardson JA, Hammer RE, Garry MG, Yanagisawa M (2005) Neuropeptide B-deficient mice demonstrate hyperalgesia in response to inflammatory pain. Proc Natl Acad Sci USA 102(28):9942–9947. https://doi.org/10.1073/pnas.0503795102
Kitamura Y, Tanaka H, Motoike T, Ishii M, Williams SC, Yanagisawa M, Sakurai T (2006) Distribution of neuropeptide W immunoreactivity and mRNA in adult rat brain. Brain Res 1093:123–134. https://doi.org/10.1016/j.brainres.2006.03.041
Lee DK, Nguyen T, Porter CA, Cheng R, George SR, O’Dowd BF (1999) Two related G protein-coupled receptors: the distribution of GPR7 in rat brain and the absence of GPR8 in rodents. Mol Brain Res 71(1):96–103
Liu M, Bu G, Wan Y, Zhang J, Mo C, Li J, Wang Y (2022) Evidence for neuropeptide W acting as a physiological corticotropin-releasing inhibitory factor in male chickens. Endocrinology. 163(7):bqac073. https://doi.org/10.1210/endocr/bqac073
Meyer A, Van de Peer Y (2005) From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27(9):937–945. https://doi.org/10.1002/bies.20293
Mondal MS, Yamaguchi H, Date Y, Shimbara T, Toshinai K, Shimomura Y, Mori M, Nakazato M (2003) A role for neuropeptide W in the regulation of feeding behavior. Endocrinology 144(11):4729–4733. https://doi.org/10.1210/en.2003-0536
Nagata-Kuroiwa R, Furutani N, Hara J, Hondo M, Ishii M, Abe T, Mieda M, Tsujino N, Motoike T, Yanagawa Y, Kuwaki T, Yamamoto M, Yanagisawa M, Sakurai T (2011) Critical role of neuropeptides B/W receptor 1 signaling in social behavior and fear memory. PLoS One 6(2):e16972. https://doi.org/10.1371/journal.pone.0016972
Price CJ, Samson WK, Ferguson AV (2009) Neuropeptide W has cell phenotype-specific effects on the excitability of different subpopulations of paraventricular nucleus neurones. J Neuroendocrinol 21(10):850–857. https://doi.org/10.1111/j.1365-2826.2009.01904.x
Sakurai T (2013) NPBWR1 and NPBWR2: implications in energy homeostasis, pain, and emotion. Front Endocrinol (lausanne) 4:23. https://doi.org/10.3389/fendo.2013.00023
Samson WK, Baker JR, Samson CK, Samson HW, Taylor MM (2004) Central neuropeptide B administration activates stress hormone secretion and stimulates feeding in male rats. J Neuroendocrinol 16(10):842–849. https://doi.org/10.1111/j.1365-2826.2004.01239.x
Shimomura Y, Harada M, Goto M, Sugo T, Matsumoto Y, Abe M, Watanabe T, Asami T, Kitada C, Mori M, Onda H, Fujino M (2002) Identification of neuropeptide W as the endogenous ligand for orphan G-protein-coupled receptors GPR7 and GPR8. J Biol Chem 277(39):35826–35832. https://doi.org/10.1074/jbc.M205337200
Singh G, Davenport AP (2006) Neuropeptide B and W: neurotransmitters in an emerging G-protein-coupled receptor system. Br J Pharmacol 148(8):1033–1041. https://doi.org/10.1038/sj.bjp.0706825
Singh G, Maguire JJ, Kuc RE, Fidock M, Davenport AP (2004) Identification and cellular localisation of NPW1 (GPR7) receptors for the novel neuropeptide W-23 by [I-125]-NPW radioligand binding and immunocytochemistry. Brain Res 1017(1–2):222–226. https://doi.org/10.1016/j.brainres.2004.03.079
Tanaka H, Yoshida T, Miyamoto N, Motoike T, Kurosu H, Shibata K, Yamanaka A, Williams SC, Richardson JA, Tsujino N, Garry MG, Lerner MR, King DS, O’Dowd BF, Sakurai T, Yanagisawa M (2003) Characterization of a family of endogenous neuropeptide ligands for the G protein-coupled receptors GPR7 and GPR8. Proc Natl Acad Sci USA 100(10):6251–6256. https://doi.org/10.1073/pnas.0837789100
Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y (2004) Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci USA 101(6):1638–1643. https://doi.org/10.1073/pnas.0307968100
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):RESEARCH0034. https://doi.org/10.1186/gb-2002-3-7-research0034
Wong AO, Zhou H, Jiang Y, Ko WK (2006) Feedback regulation of growth hormone synthesis and secretion in fish and the emerging concept of intrapituitary feedback loop. Comp Biochem Physiol A Mol Integr Physiol 144(3):284–305. https://doi.org/10.1016/j.cbpa.2005.11.021
Wu Y, He Z, Zhang L, Jiang H, Zhang W (2012) Ontogeny of immunoreactive Lh and Fsh cells in relation to early ovarian differentiation and development in protogynous hermaphroditic ricefield eel Monopterus albus. Biol Reprod 86(3):93. https://doi.org/10.1095/biolreprod.111.095646
Yamamoto T, Saito O, Shono K, Tanabe S (2005) Anti-hyperalgesic effects of intrathecally administered neuropeptide W-23, and neuropeptide B, in tests of inflammatory pain in rats. Brain Res 1045(1–2):97–106 (S0006-8993(05)00449-X)
Yang L, Sun C, Li W (2014) Neuropeptide B in Nile tilapia Oreochromis niloticus: molecular cloning and its effects on the regulation of food intake and mRNA expression of growth hormone and prolactin. Gen Comp Endocrinol 200:27–34 (S0016-6480(14)00042-2)
Yang S, Ma Z, Suo C, Cheng L, Su J, Lei Z (2018a) Cloning and mRNA expression of NPB and its effect on hormone secretion of the reproductive cells in the pig. Gen Comp Endocrinol 261:97–103 (S0016-6480(17)30225-3)
Yang W, Zhang N, Shi B, Zhang S, Zhang L, Zhang W (2018b) Isotocin regulates growth hormone but not prolactin release from the pituitary of ricefield eels. Front Endocrinol (lausanne) 9:166. https://doi.org/10.3389/fendo.2018.00166
Yang W, Zhang N, Wu Y, Zhang L, Zhang L, Zhang W (2021) Oxytocin-like signal regulates Lh cells directly but not Fsh cells in the ricefield eel Monopterus albus. Biol Reprod 104(2):399–409
Zaratin PF, Quattrini A, Previtali SC, Comi G, Hervieu G, Scheideler MA (2005) Schwann cell overexpression of the GPR7 receptor in inflammatory and painful neuropathies. Mol Cell Neurosci 28(1):55–63. https://doi.org/10.1016/j.mcn.2004.08.010
Acknowledgements
The authors would like to thank Xi Li and Riping Gan for their help in collecting tissue samples of ricefield eels.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32072956 and 31572604).
Author information
Authors and Affiliations
Contributions
All authors meet the authorship requirements. W. Z., L. Z., and W. Y. conceived the study, analyzed data, and wrote the manuscript. W. Y., S. Q., and X. L. contributed in acquisition of data. W. Z. and L. Z. contributed in acquisition of funding. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
All procedures and investigations were reviewed and approved by the Center for Laboratory Animals of Sun Yat-sen University and were performed in accordance with the Guiding Principles for the care and use of laboratory animals.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, W., Qian, S., Li, X. et al. Neuropeptide B (NPB) and NPB receptor 2b (NPBWR2b) in the ricefield eel Monopterus albus: expression and potential involvement in the regulation of gonadotropins. Fish Physiol Biochem 49, 983–1003 (2023). https://doi.org/10.1007/s10695-023-01237-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-023-01237-x