Abstract
Humic substances, a major component of natural organic matter in surface waters, can induce biotransformation enzyme activities and influence antioxidant defense in fish. The study aimed to provide a molecular basis for the stress responses, the synthesis of biotransformation, and antioxidant enzymes in particular. Adult medaka fish (Hd-rR strain) were exposed to environmentally relevant concentrations of humic acid for 96 h. The actual humic acid concentrations in water were determined photometrically and expressed as organic carbon concentrations. Liquid chromatography with tandem mass spectrometry was used for protein profile analysis of medaka liver samples. The relative amount of isozymes was determined using the label-free quantification approach. Hepatic biotransformation enzyme activities were measured as well. Thus, ethoxyresorufin-O-deethylase activity showed a pronounced induction at the highest tested concentration (9.4 mg C/L). Various biotransformation and antioxidant isozymes responded to humic acid differently, reflecting a balanced interplay of proteins that ensures the metabolism of humic acid in fish liver. Some isozymes were not affected by humic acid. The study provides new insight into the molecular mechanisms of the fish stress response to the humic acid-related challenge.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Humic substances are major components of natural organic matter in surface waters and, at higher concentrations, can impart a dark color, especially in brown-colored freshwaters. Humic substances are complex mixtures formed by biochemical and chemical reactions during the decay and transformation of plant and microbial remains. Aquatic humic substances operationally can be separated into humic acid (HA) and fulvic acid fractions (MacCarthy 2001; IHSS, 2007; Olk et al. 2019). Depending on the analytical approach, the amount of humic substances in natural waters can be expressed in organic carbon concentrations (mg C/L) or the total amount of humic substances (mg/L). The lowest organic carbon concentrations were found in sea and groundwater: 0.5 and 0.7 mg C/L, respectively (Thurman 1985). In surface waters, organic carbon concentrations varied significantly, from around 2 mg C/L in oligotrophic lakes to 7 mg C/L in upland rivers and 12–14 mg C/L in eutrophic lakes (Thurman 1985; Wiegand et al. 2007; Rodríguez and Núñez 2011), up to 41–61 mg C/L in rivers fed by wetlands (Kihara et al. 2012). When expressed as the total amount of humic substances extracted from water, Linnik et al. (2013) reported the range of 1.2 to 48 mg/L humic substances in rivers and reservoirs up to 126.5 mg/L in some parts of rivers fed by wetlands.
There is no doubt that humic substances are an ecological driving force in freshwater ecosystems due to their ability to influence aquatic organisms, populations, and communities (reviewed in Steinberg et al. 2006, 2009). Humic substances are able to permeate animal cells (Beer et al. 2003). Humic substances have been found to cause various effects in aquatic organisms, including reducing photosynthetic oxygen production, chemical attraction, estrogenicity, physical and chemical membrane irritation, induction and modulation of biotransformation enzymes, induction of stress defense proteins, or oxidative stress defense (reviewed in Steinberg et al. 2003, 2009).
Of particular interest for us is the ability of pure humic substances to induce biotransformation enzyme activities (Steinberg et al. 2003; Bittner et al. 2006; Matsuo et al. 2006) and influence the antioxidant status in fish (Adekunle and Ajuwon 2010; Alak et al. 2013; Deng et al. 2020). If the acclimatization potential of fish is overwhelmed by external factors, an organism may enter stress conditions. The most studied environmental stress conditions in aquatic environments include changes in salinity, ion composition, temperature, and oxygen availability, as well as exposure to pollutants due to natural processes or human activity (Lushchak 2016). Fish responses to humic substance-related challenge are far less understood.
To perform this study, we chose to use both catalytic and protein profiling approaches. Mass-spectrometry-based protein profiling has become an increasingly powerful technology for the identification of proteins and their quantification. Label-free quantification (LFQ) approach using the MaxLFQ algorithms, part of the MaxQuant software suite, can achieve the highest possible quantification accuracy (Cox et al. 2014) and provide data on the relative amount of proteins in two or more biological samples. The objectives of our study were to evaluate the effect of humic substances on the ethoxyresorufin-O-deethylase (EROD) and glutathione-S-transferase (GST) catalytic activities, well-established biomarkers of xenobiotic exposure, and the relative amounts of biotransformation and antioxidant proteins, cytochromes P450, glutathione-S-transferase, UDP-glucuronosyltransferase, superoxide dismutase, catalase, glutathione peroxidase, and peroxiredoxin, in the liver of medaka exposed to environmentally relevant HA concentrations.
Medaka or Japanese rice fish (Oryzias latipes) is a small (3–4 cm) freshwater fish that have been used extensively in various experimental studies (Lin et al. 2014). Medaka exhibits morphological dimorphism between males and females and is easy to rear in the laboratory; its genome has been completely sequenced (Kasahara et al. 2007), and a reference proteome is provided (UniProt, 2021a). These characteristics make medaka a valuable model organism.
Materials and methods
Fish, exposure, and sampling
Medaka’s fertilized eggs (Hd-rR strain) were initially obtained through the USGS CERC – IBIW RAS Scientific Program. Medaka has been maintained in the Laboratory of Physiology and Toxicology of Aquatic Animals (IBIW RAS) for 5 years. Medaka breeding stock was kept in a flow-through system at 25 ± 1 °C and 16:8 h photoperiod in water similar to the test water. Tap water from the groundwater source was used (conductivity 462 ± 6 µS/cm, pH 7.97–8.12) with mechanical aeration provided by air-delivery lines. Prior to HA exposure, a stock intended for this study was held at the same temperature and photoperiod regime under semi-static conditions (50% daily water renewal) for a month. The loading was kept at ≤ 1 g wet weight of fish per liter.
Adult medaka (40–45 weeks old, Table 1) were exposed to environmentally relevant concentrations (0, 5, 40, and 80 mg/L, nominal; for measured concentrations, see Table 1) of HA (CAS 1415–93-6, Sigma-Aldrich) for 96 h under semi-static conditions. After the test solutions were made, the pH was measured (Table 1). Since the values were close and fell into the recommended range of pH 6.0–8.5, only one control group (dilution water control) was used, and no additional control group (with the adjusted pH) was involved in the testing. During the exposure period, pH and dissolved oxygen values in test chambers were measured daily in duplicate (Table 1).
On day 0 of the experiment, testing solutions were prepared by dissolving the needed amount of dry HA in 10 L of water. For example, 0.8 g of HA was triturated in a bowl, dissolved in a small volume of water, and transferred into a graded tank. The final volume was brought up to10 L to obtain an 80 mg/L testing solution. Testing solutions for renewal were made daily by dilution of an 80 mg/L solution prepared as described above. According to the product specification sheet, the Sigma-Aldrich HA is soluble in water (0.1 g in 10 mL) and turbid (i.e., contains insoluble matter in a solution). The physical and chemical properties of Sigma-Aldrich HA were studied earlier (Valencia et al. 2013).
Test chambers (one per treatment) were filled with 10 L of the solutions, and the loading was ≤ 0.5 g/L (OECD 2019; Table 1). Although the main idea is that the fish should be randomly distributed among treatments, we kept a count of males and females placed in each chamber to achieve an equal proportion of the sexes.
Fish were fed commercial food (TetraMin Mini Granules) four times per day using automatic feeders and Artemia nauplii twice a day manually. Residual food granules and feces were removed daily during the water renewal. Observations for any clinical signs in fish were performed at least twice a day (OECD 2019); medaka was observed for loss of equilibrium, abnormal swimming behavior, and abnormal ventilatory function.
At the end of the exposure period, after anesthetization with tricaine methanesulfonate (MS-222, 100 mg/L), the fish was weighed, observed for visible abnormalities: exophthalmia, hemorrhage, or abnormal skin pigmentation. The liver was excised on ice, weighed, and placed into a 200-µL centrifuge tube. Ice-cold phosphate-buffered saline was added in a 1:3 ratio (w/v), and a sample was then homogenized with a pestle for 3 min on ice. Homogenate aliquots from 8 individual liver samples (from 4 males and 4 females) in each treatment group were pooled and frozen in liquid nitrogen to be used later for the protein profiling assay. The bulk of the individual liver homogenates were then centrifuged at 12000 g for 15 min at 4 °C to be used for catalytic enzyme activity assays. The resulting supernatants were collected, avoiding the lipid phase, and stored in liquid nitrogen until analyzed.
Organic carbon analysis
In semi-static renewal experiments, test concentrations should be measured at least twice over one exposure period (before and after the renewal of test solutions) (OECD 2019). Therefore, we sampled test solutions at 0 h (before distributing fish among treatments) and 78 h (6 h after the third renewal of test solutions). HA solutions were analyzed in duplicate. Water samples were centrifuged at 10000 g for 5 min to separate the non-dissolved and any other interfering particles, and supernatants were used for the assay. Organic carbon concentrations were measured employing the potassium dichromate photometric method using a KFK-3 instrument (ZOMZ, Russia) (Shamrikova et al. 2012).
Ethoxyresorufin-O-deethylase activity assay
EROD activity was determined by measuring the concentration of resorufin (Kennedy and Jones 1994) normalized by protein concentration (see the “Total protein assay” section) in supernatants using 96-well plates. To each well, 50 μL of 100 mM potassium phosphate-buffered saline (pH 7.5), 10 μL supernatant aliquot, 50 μL of 10 μM 7-ethoxyresorufin (in methanol), and 50 μL of 4.3 μM β-NADPH tetrasodium salt were added. A well plate was incubated for 10 min at 27 °C using an ST-3 Sky Line thermostatic shaker (ELMI, Latvia). Fluorescence of the resorufin produced was measured at excitation/emission wavelengths of 555/585 nm in an LS 55 fluorescence spectrometer (PerkinElmer, UK). The amount of resorufin produced was calculated using a resorufin standard curve plotted using a commercially available product (CAS 635–78-9, MP Biomedicals, USA). Each sample was measured in triplicate and reported in pM/mg protein/min.
Glutathione-S-transferase activity assay
GST activity was determined by monitoring the conjugation of reduced glutathione with 1-chloro-2,4-dinitrobenzene used as a substrate (Habig et al. 1974), normalized by protein concentration. Analysis was carried out in 96-well plates. The reaction mixture was composed of 11.640 mL of 100 mM potassium phosphate-buffered saline (pH 6.5), 240 μL of 100 mM reduced glutathione, and 120 μL of 100 mM 1-chloro-2,4-dinitrobenzene. A 2 μL supernatant aliquot was pipetted to each well, and 98 μL of the reaction mixture was added. The increase in absorbance was measured at 340 nm and recorded for 3 min using a SPECTROstar Nano spectrometer (BMG LAB-TECH, Germany). Each sample was measured in triplicate and reported in µM/mg protein/min.
Total protein assay
Total protein content in supernatant samples was measured using a fluorescamine reaction (Udenfriend et al. 1972; Lorenzen and Kennedy 1993) with the bovine serum albumin used as a standard in 96-well plates. A series of bovine serum albumin dilutions ranging from 0 to 4.8 mg/mL was made in 100 mM potassium phosphate-buffered saline (pH 7.5). Fluorescamine was dissolved in acetone to obtain 10.8 mM solution (3 mg/mL). Protein analysis was carried out immediately after the EROD assay in the same microplate. To each well, 150 µL of phosphate-buffered saline and 50 µL of fluorescamine solution were added. The plate was shaken for 1 min in an ELMI ST-3 M microplate shaker. The fluorescence was then determined at 400/460 nm excitation/emission wavelengths using an LS 55 fluorescence spectrometer (PerkinElmer).
Liquid chromatography-mass spectrometry protein analysis
S-Trap Mini Spin Columns (ProtiFi) were used for trypsin protein digestion according to the manufacturer’s instructions (S-Trap™ micro spin column digestion protocol 4.7). Further analysis of obtained peptides was performed by high-performance liquid chromatography with tandem mass spectrometry (HPLC–MS/MS) described below. A 1-µg aliquot of peptides in a volume of 1–4 µL was loaded onto the Acclaim µ-Precolumn (0.5 × 3 mm, 5 µm particle size, Thermo Scientific) at a flow rate of 10 µL/min for 4 min in an isocratic mode of Mobile Phase C (2% acetonitrile, 0.1% formic acid). Then, the peptides were separated with HPLC (Thermo Scientific™ UltiMate™ 3000 RSLCnano system, Rockwell, IL, USA) in a 15-cm long C18 column (Acclaim® Pep-Map™ RSLC inner diameter of 75 μm, Thermo Fisher Scientific, Rockwell, IL, USA). The peptides were eluted with a gradient of buffer B (80% acetonitrile, 0.1% formic acid) at a flow rate of 0.3 μL/min. Total run time was 90 min, which included initial 4 min column equilibration to buffer A (0.1% formic acid), a gradient from 5 to 35% of buffer B for 65 min, then 6 min to reach 99% of buffer B, 10 min flushing with 99% of buffer B, and 5 min re-equilibration to buffer A. The MS analysis was performed in triplicate using a Thermo Scientific™ Q Exactive HF-X hybrid quadrupole-Orbitrap mass spectrometer (Rockwell, IL, USA). The temperature of the capillary was 240 °C, and the voltage at the emitter was 2.1 kV. Mass spectra were acquired at a 120,000 resolution in a range of 300–1500 m/z. Tandem mass spectra of fragments were acquired at a 15,000 resolution in a range from 100 m/z to the value determined by a charge state of the precursor, but not higher than 2000 m/z. The maximum integration time was 50 ms/110 ms for precursor and fragment ions, correspondingly. AGC (automatic gain control) target values for precursor and fragment ions were 1E6 and 2E5, correspondingly. An isolation intensity threshold of 50,000 counts was determined for precursor selection, and up to the top 20 precursors were chosen for fragmentation with HCD (higher-energy collisional dissociation) at normalized collision energy (NCE) of 29. Precursors with a charge state of 1 + and more than 5 + were rejected, and all measured precursors were dynamically excluded from triggering a subsequent MS/MS for 20 s.
Obtained raw data were processed using the MaxQuant software (version 1.6.3.4) with the built-in Andromeda peptide search engine. Protein sequences for the Oryzias latipes provided by UniProt (Feb 2021) were used for protein identification. Carbamidomethylation of cysteines was set as fixed modification, and oxidation of methionines was set as a variable modification for the peptide search. A maximum m/z deviation of 4.5 ppm was allowed for precursor identification, and 20 ppm was set as match tolerance for fragment identification (acquisition in Orbitrap). One missed cleavage was allowed for trypsin digestion. The software option “Match between runs” was enabled, and features within a time window of 2 min were used to match between runs. The false discovery rates for peptide and protein identifications were set to 5%. Only proteins with minimally two peptides detected were considered reliably identified.
Statistical analysis
In HPLC–MS/MS protein quantification analysis, three technical replicates of each pooled homogenate sample were used in HPLC–MS/MS protein quantification analysis. LFQ intensity values from these replicates were log2-transformed, and ANOVA with post hoc Fisher’s LSD was used to identify differences among samples (= treatments). Pearson correlation coefficients (R) were determined for log2-transformed LFQ values to evaluate the strength of the association among various proteins. EROD and GST catalytic activities were measured in the individual samples. Enzyme activity data were tested for normality using the Shapiro–Wilk test. Since the data did not meet the assumption of normality, nonparametric methods, the Kruskal–Wallis test, followed by the Mann–Whitney U test for pairwise comparison, were used to determine statistically significant differences among the treatments. Spearman rank correlation coefficient (RS) was used to determine the associations between EROD or GST activity and organic carbon concentrations in the water.
Results
Results on measured organic carbon concentrations in the testing solutions are given in Table 1.
No mortalities were registered during the exposure period in any treatment group. No clinical signs of toxicity were observed either: no loss of equilibrium, abnormal swimming behavior (hypo-, hyperactivity, corkscrew swimming, convulsions, paralysis, loss of/ dense schooling, over-/under-reaction to stimulus), or abnormal ventilatory function (hyper-/hypo-/irregular ventilation, “coughing,” gulping). However, in treatments 2 and 3, the coloring of water reduced the visibility; therefore, intravital clinical signs could not be adequately examined and cannot be eliminated with certainty. During sampling, no visible exterior abnormalities (exophthalmia, hemorrhage, or abnormal skin pigmentation) in medaka were found.
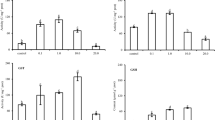
There were no statistically significant differences in EROD and GST activities between males and females in each treatment group (Mann–Whitney U test, p > 0.05). Hepatic EROD activity values varied widely within treatment groups by the end of 96-h exposure (Fig. 1a). A statistically significant increase in activity was evident in treatment 3. Median values of GST activity in treatment groups were lower than that in control (Fig. 1b).
The relative amounts of HPLC–MS/MS identified biotransformation and antioxidant proteins are reported as log2-transformed LFQ values and presented in Fig. 2. Three cytochrome P450 isozymes were detected in medaka liver samples (Fig. 2a). CYP1A1 was the least abundant. Log2(LFQ) values for CYP1A1 were reported in all treatments, but one (zero LFQ values were registered in all replications in treatment 1). CYP1A1 did not differ significantly between treatment 2 and the control but was significantly upregulated in treatment 3. CYP2P3 showed the difference between treatments 2 and 3. No significant variability among treatments was registered for CYP3A40. Correlation analysis showed no considerable correlation among the cytochrome P450 enzymes (Online Resource 1).
The relative amount of proteins expressed in the log2-transformed LFQ values. Plots are constructed using mean ± SD values. Letters denote statistically significant differences among treatment groups (ANOVA with post hoc Fisher’s LSD, p < 0.05). In plot (d), an asterisk marks a column constructed from a single log2(LFQ) value
Six GST isozymes were identified in medaka liver samples (Fig. 2b). The most abundant were GSTA4, MGST1, and GSTM4. The greatest amount of GSTA4 was registered in treatment 1, the smallest in treatment 2. No significant differences between treatments were found in the amount of MGST1. GSTM4 showed a maximum level in treatment 1 and decreased gradually in treatments 2 and 3. No significant differences were observed in log2(LFQ) means of GST (LOC101157367). With no changes in treatments 1 and 2, the amount of GST (LOC101170897) was significantly greater in treatment 3. The most pronounced differences were seen in GST (LOC101173040); the least abundant in control, it was dramatically induced in all treatments. A strong negative correlation was found between two GST isozymes, GST (LOC101173040) and GST (LOC101157367), R = − 0.85, p = 0.02.
Five UDP-glucuronosyltransferase (UGT) isozymes were detected in the medaka liver, showing different relative abundance patterns (Fig. 2c). UGT (LOC100125517), the most abundant in the analyzed samples, had gradually decreased log2(LFQ) values. A significant increase in protein amount was registered in treatment 1, and a decrease in treatment 3. UGT (LOC101168388) and UGT (LOC101159946) were slightly upregulated in treatment 1 and significantly downregulated in treatment 2. UGT (N/A) isozyme had the greatest amount in control, and showed a slight decrease in treatment 1 and significantly smaller amounts in treatments 2 and 3. The least abundant in the samples, UGT (LOC101173361), showed no significant differences among treatments; its pattern resembled that of UGT (N/A). Strong positive correlations were found among three UGT isozymes, UGT (LOC101168388) and UGT (N/A), R = 0.91, p = 0.004, UGT (LOC101168388) and UGT (LOC101159946), R = 0.94, p = 0.002, UGT (LOC101159946) and UGT (N/A), R = 0.78, p = 0.04.
Three superoxide dismutase isozymes were detected in medaka liver samples (Fig. 2c). SOD1 was the most abundant isozyme; it was downregulated in treatments 1 and 2. SOD2 showed a close pattern but was characterized by more pronounced upregulation in treatment 3. SOD3 had a reverse distribution among the samples: the greatest amount of the protein was registered in treatment 1, while in treatment 3, it was detected in one out of three replicates. A strong positive correlation was found between SOD1 and SOD2, R = 0.98, p = 0.0001, and negative between SOD2 and SOD3, R = − 0.77, p = 0.04.
The relative amounts of catalase and glutathione peroxidase, GPX1, are presented in Fig. 2e. In its distribution among treatments, catalase showed a close resemblance with SOD1, but with more pronounced graded increases from treatment 1 to treatment 3. The amounts of glutathione peroxidase were very close in control and treatments 1 and 2 and decreased in treatment 3.
Three peroxiredoxin isozymes were detected in the samples (Fig. 2f), showing very similar patterns and varying only in the abundance and the extent of up-/downregulation. Strong positive correlations were found between PRDX2 and peroxiredoxin-1 (LOC101175597), R = 0.85, p = 0.01, and PRDX2 and PRDX5, R = 0.85, p = 0.02.
Results of the correlation analysis performed on the log2(LFQ) values of the detected biotransformation and antioxidant proteins are given in Online Resource 1. Strong correlations were found among the studied enzymes, except for CYP3A40 and MGST1.
Discussion
Results of organic carbon analysis lacked the resemblance with nominal concentrations of HA in testing solutions. This fact can be explained by the physical–chemical properties of the HA product and its behavior in dilution water (presence of insoluble matter in a solution). Still, the measured concentrations of organic carbon dissolved in the water were within the range typical for aquatic ecosystems (Thurman 1985; Wiegand et al. 2007; Rodríguez and Núñez 2011).
One of the documented humic substances effects in fish is the induction of biotransformation proteins such as cytochrome P450 1A (CYP1A) (Matsuo et al. 2006; Zhao et al. 2017). Biotransformation is a process catalyzed primarily through enzymatic reactions that often alter the chemistry of nonpolar lipophilic chemicals to polar water-soluble metabolites, leading to the elimination of the parent compounds. When polarity has been enhanced through phase I reactions, phase II reactions generally attempt to further enhance polarity by conjugating the phase I product with a bulky polar endogenous molecule (Schlenk et al. 2008). Cytochromes P450 have a broad spectrum of substrates and are essential in phase I biotransformation reactions. These heme proteins operate by binding and activating molecular oxygen and thereby oxidizing the substrate (Coppock and Dziwenka 2014). Providing evidence of the CYP1A upregulation by xenobiotic chemicals, EROD induction is a highly sensitive biomarker of contaminant exposure in fish (Whyte et al. 2000). Our results showed a well-pronounced increase in EROD activity at the highest tested HA concentration (80 mg/L, nominal). In a similar experiment with synthetic HA (Andersson et al. 2010), resembling effects were observed in the liver of female three-spined sticklebacks (Gasterosteus aculeatus) exposed for 48 h. Weak EROD induction was registered in fish exposed to 20 mg/L, while at 100 mg/L, a significant increase was seen compared with the control. Interestingly, at the same time, HA caused strong concentration-dependent EROD induction in the gills of the three-spined sticklebacks, thus providing evidence of CYP1A upregulation in response to HA exposure (Andersson et al. 2010).
Unexpectedly, considering the results of hepatic EROD activity in medaka, the HPLC–MS/MS analysis did not show any CYP1A1 protein in the pooled medaka liver sample in treatment 1. However, clear evidence of CYP1A1 upregulation was seen at the highest tested HA concentration. As for CYP2P3 and CYP3A40, their response to HA was far less pronounced, even though they are known to participate in the metabolism of various xenobiotics (Oleksiak et al. 2003; Tseng et al. 2005; Zhang et al. 2017). These cytochrome P450 enzymes are monooxygenases, i.e., catalyze the incorporation of one atom from molecular oxygen into a compound and reduction of the other atom of oxygen to water, and act on a wide variety of organic substrates. CYP1A1 and CYP3A40 are endoplasmic reticulum membrane-associated proteins (UniProt 2021c, 2021j); CYP2P3 is located in the endoplasmic reticulum membrane and cytoplasm (UniProt 2021e). In this study, CYP2P3 showed a weak response to HA exposure, and it seemed that CYP3A40 was not affected by HA. At the same time, CYP1A1, the least abundant in the liver, was the most responsive to HA exposure at the highest tested concentration.
CYP1A1 is regulated by the aryl hydrocarbon receptor (AhR). It was reported that HAs are able to activate AhR-mediated pathways (Bittner et al. 2006; Janošek et al. 2007). The authors hypothesized that small molecules released from humic substances are likely to bind to the ligand-binding region of the AhR, being weak activators when compared to “classical” AhR ligands (polycyclic aromatic and halogenated hydrocarbons). Cytochromes P450 3A gene expression is regulated by the pregnane X receptor (PXR) (Gao et al. 2007). The ligand pockets of PXR are lined by mostly hydrophobic residues, and polar groups can destabilize interactions in the hydrophobic areas of the PXR ligand-binding pocket (Gao et al. 2007; Pavek 2016). Being amphiphilic compounds, HA have polar functional groups in their structures (Gomes de Melo et al. 2016). Thus, HA might not be an effective PXR activator, and therefore, no hepatic CYP3A40 upregulation in fish was registered in this study. Another possible explanation would be PXR cross-talks with other nuclear receptors and competition for coactivators (Pavek 2016).
GST are involved in the phase II metabolism of a wide variety of xenobiotics and endogenous compounds (Gertsch 2007). As opposed to the expected induction of catalytic activity, our results showed that HA did not cause any increase in GST activity in the liver of adult medaka. Moreover, HA slightly inhibited GST activity in all treatments. It was reported by Cazenave et al. (2006) that natural organic matter also caused a decrease in GST activity in embryos of zebrafish (Danio rerio) exposed for 24 h.
The HPLC–MS/MS analysis showed that GST isozymes were expressed differently in response to HA. The most responsive to HA stimulus was GST (LOC101173040), the least abundant in control. Interestingly, no concentration-dependent induction of GST (LOC101173040) was evident; the relative amount of this isozyme was equal at both minimum and maximum HA concentrations. One of the most abundant isozymes, GSTA4, had no apparent response to HA, neither did MGST1 and GST (LOC101157367). GSTM4 was upregulated at the lowest tested HA concentration, while GST (LOC101170897) showed no response to low and medium concentrations and was upregulated only at the highest. It was reported previously by Pflugmacher et al. (2001) that low humic substance concentrations showed ambiguous effects on the catalytic activity of microsomal and soluble GST in freshwater invertebrates, which can indirectly confirm our findings that GST isozymes show different responses to HA.
GST have multiple biological roles, including cell protection against oxidative stress and toxic molecules, and are involved in synthesizing and modifying leukotrienes and prostaglandins. GST are able to conjugate glutathione to a wide range of hydrophobic and electrophilic molecules, making them less toxic and predisposed to further modification for discharge from the cell. They can exhibit glutathione peroxidase activity and catalyze the reduction of organic hydroperoxides to their corresponding alcohols. GST are also implicated as modulators in signal transduction pathways (Allocati et al. 2018). HA is a complex and heterogeneous mixture of polydispersed materials; therefore, it is explicable that inside cells, HA can intervene with multiple metabolic pathways and modulate the regulation of various enzymes.
UGT are the endoplasmic reticulum-associated enzymes that catalyze the attachment of a glucuronic acid moiety to various compounds, endogenous and xenobiotic, in phase II metabolism. Glucuronic acid conjugation to the hydroxyl, carboxyl, amino, or sulfhydryl group of a target compound promotes its excretion (Meech et al., 2018). Phase II isozymes are known to be coordinately induced with cytochromes P450, phase I enzymes. Our results illustrate such co-induction of the biotransformation enzymes in response to HA exposure. Thus, CYP1A1 was strongly associated with GST (LOC101170897), CYP2P3 with UGT (LOC101159946), and UGT (LOC101168388). Several phase II enzymes showed “unidirectional” responses: GSTA4 with UGT (N/A) and UGT (LOC101173361); GST (LOC101157367) with UGT (N/A) and UGT (LOC101168388). At the same time, certain enzymes had strong inverse relationships: CYP1A1 with GSTM4 and UGT (LOC100125517), GST (LOC101173040) with GST (LOC101157367), UGT (N/A), UGT (LOC101168388), and UGT (LOC101159946), reflecting the balance of up- and downregulation, maintaining the metabolism of HA.
Many studies have found that environmental challenges can alter redox homeostasis in fish. Like other living organisms, fish possess multilevel and complicated antioxidant systems operating to prevent reactive oxygen species formation, eliminate reactive oxygen species and reactive oxygen species-modified molecules, or minimize their harmful effects (reviewed in Lushchak 2016). SOD are a group of metalloenzymes (requiring a metal cofactor for activity) that form the front line of defense against reactive oxygen species-mediated injury. SOD catalyze the dismutation of superoxide anion radicals into molecular oxygen and hydrogen peroxide (Younus 2018). Copper, zinc-dependent SOD1 was the most abundant in the liver samples of medaka. This isozyme is located in the cytosol, mitochondria, nucleus, and peroxisome (UniProt 2021d). HA did not seem to affect copper, zinc-dependent SOD1, even though it is known to respond to xenobiotic stimulus. Manganese-dependent SOD2, localized solely in the mitochondrion matrix (UniProt 2021h), showed a similar response. The third identified SOD isozyme, copper, zinc-dependent SOD3, is an extracellular enzyme (InterPro 2021); its function is to protect the extracellular space from stress by oxidants (Nozik-Grayck et al. 2005). SOD3 showed reversed expression pattern. Taken together, these results indicate that the detected SOD isozymes, to a certain extent, compensate for each other’s action in response to HA impact in the cell and extracellular space.
Catalase is responsible for reducing hydrogen peroxide to water and molecular oxygen; it is principally limited to the peroxisomes. Glutathione peroxidase, a selenium-dependent enzyme located in cytosol and mitochondrion, is a crucial antioxidant enzyme involved in the breakdown of hydrogen peroxide (Lubos et al. 2011; UniProt 2021f). In medaka liver samples, these enzymes showed a close inverse relationship. Peroxiredoxins are thiol-specific peroxidases that also catalyze the reduction of hydrogen peroxide and organic hydroperoxides. Peroxiredoxin-1 isozymes are located in the cytosol (UniProt 2021g, i), and peroxiredoxin-5 is attributed to the peroxisome matrix (UniProt 2021b). Hydrogen peroxide is the product of SOD catalytic activity; therefore, it was expected to find strong correlations between SOD isozymes and catalase, glutathione peroxidase, and peroxiredoxins. Such close associations were found between catalase and copper, zinc-dependent SOD1, and manganese-dependent SOD2, and between glutathione peroxidase and peroxiredoxin-1 (LOC101175597). At the same time, inverse relationships were observed between catalase and copper, zinc-dependent SOD3, glutathione peroxidase, peroxiredoxin-1 (prdx2), and peroxiredoxin-1 (LOC101175597), as well as both peroxiredoxin-1 isozymes with SOD1 and SOD2 or glutathione peroxidase with SOD2. Catalase, glutathione peroxidase, and peroxiredoxin play their parts in cell protection against oxidative stress by detoxifying peroxides. Besides, antioxidant enzymes are involved in redox signaling, and different isozymes may be linked to different signaling pathways (Forman et al. 2014; Rhee 2016). These results illustrate a balanced interplay of antioxidant isozymes in the liver of medaka in response to HA exposure.
In teleost fish, xenobiotic-metabolizing enzymes are typically modulated through the activation of nuclear receptors (ligand-regulated transcription factors), controlling induction of cytochromes, as well as other phase I oxidative enzymes, phase II conjugating enzymes, and phase III transport uptake and efflux transporters, coordinately regulating xenobiotic clearance in the liver (Orans et al. 2005; Ma 2011; Saad et al. 2016). Xenobiotics can also act as prooxidant stressors, increasing the intracellular generation of reactive oxygen species, which modulate levels and functions of redox-sensitive signaling proteins and antioxidants. A network of antioxidant and cytoprotective enzymes, including copper, zinc-dependent SOD, catalase, glutathione peroxidase, peroxiredoxin, GST, and UGT, is modulated by the nuclear factor erythroid 2-related factor (Nrf2) (Regoli et al. 2011; Loboda et al. 2016). Nrf2 is a transcription factor that coordinates the basal and stress-inducible activation of a vast array of cytoprotective genes and therefore represents a crucial regulator of the cellular defense mechanisms against xenobiotic and oxidative stress (Tonelli et al. 2018). Therefore, it is expected to observe concordance among biotransformation and antioxidant enzymes caused by the need to eliminate HA, provide antioxidant defense against its potential detrimental effects, and maintain basal metabolic processes. In our study, relative amounts of CYP1A1 had strong relationships with manganese-dependent SOD2 and catalase, and strong inverse relationships with copper, zinc-dependent SOD3, glutathione peroxidase, and both peroxiredoxin-1 enzymes. CYP2P3 showed strong associations with manganese-dependent SOD2, moderate association with copper, zinc-dependent SOD1, and a strong inverse relationship with peroxiredoxin-1 (LOC101175597). The results as a whole suggest that environmentally relevant HA concentrations evoke a coordinated response of the hepatic biotransformation and antioxidant defense systems in fish. Some proteins were upregulated at the lowest nominal concentration (e.g., UGT (LOC100125517), SOD3, and PRDX2); the others were at the highest (CYP1A1, GST (LOC101170897), SOD2, and catalase). GST (LOC101173040) was affected by HA presence the most, while CYP3A40 and GST (LOC101157367) showed no clear response to this challenge.
Conclusions
Short-term exposure to environmentally relevant concentrations of HA caused catalytic and isozyme abundance changes in biotransformation and antioxidant defense systems in the liver of adult medaka fish. Further research addressing prolonged stress responses would help gain new knowledge of molecular mechanisms that allow fish to cope with the persisting environmental humic substances-related challenge.
Availability of data and material
Raw data generated during the current study are available from the corresponding author on reasonable request. Correlation analysis data is available in Supplementary Information.
References
Adekunle IM, Ajuwon OR (2010) Influence of humic acid derived from composted wastes of Nigeria origin on oxidative and antioxidant status of African mud catfish (Clarias gariepinus). Pak J Biol Sci 13:821–827
Alak G, Atamanalp M, Topal A, Arslan H, Oruç E, Altun S (2013) Histopathological and biochemical effects of humic acid against cadmium toxicity in brown trout gills and muscles. Turk J Fish Aquat Sc 13:315–320. https://doi.org/10.4194/1303-2712-v13_2_13
Allocati N, Masulli M, Di Ilio C, Federici L (2018) Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 7:1–15. https://doi.org/10.1038/s41389-017-0025-3
Andersson C, Abrahamson A, Brunström B, Örberg J (2010) Impact of humic substances on EROD activity in gill and liver of three-spined sticklebacks (Gasterosteus aculeatus). Chemosphere 81:156–160. https://doi.org/10.1016/j.chemosphere.2010.06.073
Beer A-M, Junginger HE, Lukanov J, Sagorchev P (2003) Evaluation of the permeation of peat substances through human skin in vitro. Int J Pharm 253:169–175. https://doi.org/10.1016/S0378-5173(02)00706-8
Bittner M, Janošek J, Hilscherová K, Giesy J, Holoubek I, Bláha L (2006) Activation of Ah receptor by pure humic acids. Environ Toxicol 21:338–342. https://doi.org/10.1002/tox.20185
Cazenave J, Bistoni MDLÁ, Zwirnmann E, Wunderlin DA, Wiegand C (2006) Attenuating effects of natural organic matter on microcystin toxicity in zebra fish (Danio rerio) embryos – benefits and costs of microcystin detoxication. Environ Toxicol 21:22–32. https://doi.org/10.1002/tox.20151
Coppock RW, Dziwenka MM (2014) Biomarkers of petroleum products toxicity. In: Gupta RC (ed) Biomarkers in Toxicology. Academic Press, pp 647–654. https://doi.org/10.1016/B978-0-12-404630-6.00037-3
Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalisation and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. https://doi.org/10.1074/mcp.M113.031591
Deng J, Lin B, Zhang X, Guo L, Chen L, Li G, Wang Q, Yu C, Mi H (2020) Effects of dietary sodium humate on growth, antioxidant capacity, non-specific immune response, and resistance to Aeromonas hydrophila in genetic improvement of farmed tilapia (GIFT, Oreochromis niloticus). Aquaculture 520:734788. https://doi.org/10.1016/j.aquaculture.2019.734788
Forman HJ, Ursini F, Maiorino M (2014) An overview of mechanisms of redox signalling. J Mol Cell Cardiol 73:2–9. https://doi.org/10.1016/j.yjmcc.2014.01.018
Gao Y-D, Olson SH, Balkovec JM, Zhu Y, Royo I, Yabut J, Evers R, Tan EY, Tang W, Hartley DP, Mosley RT (2007) Attenuating pregnane X receptor (PXR) activation: a molecular modelling approach. Xenobiotica 37:124–138. https://doi.org/10.1080/00498250601050412
Gertsch J (2007) Glutathione-S-transferase. In: Enna SJ, Bylund DB (eds) xPharm: The Comprehensive Pharmacology Reference. Elsevier, pp 1–17. https://doi.org/10.1016/B978-008055232-3.60523-9
Gomes de Melo BA, Lopes Motta F, Andrade Santana MH (2016) Humic acids: structural properties and multiple functionalities for novel technological developments. Mat Sci Eng C-Mater 62:967–974. https://doi.org/10.1016/j.msec.2015.12.001
Habig WH, Pabst MJ, Jakoby WB (1974) Glutathione S-transferases: the first enzymatic step in mercapturic acid formation. J Biol Chem 249:7130–7139. https://doi.org/10.1016/S0021-9258(19)42083-8
IHSS (2007) International Humic Substances Society. https://humic-substances.org/what-are-humic-substances-2/. Accessed on 14 March 2021
InterPro (2021) Extracellular superoxide dismutase [Cu-Zn]. https://www.ebi.ac.uk/interpro/entry/InterPro/IPR024141/. Accessed on 26 April 2021
Janošek J, Bittner M, Hilscherová K, Bláha L, Giesy JP, Holoubek I (2007) AhR-mediated and antiestrogenic activity of humic substances. Chemosphere 67:1096–1101. https://doi.org/10.1016/j.chemosphere.2006.11.045
Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y (2007) The medaka draft genome and in-sights into vertebrate genome evolution. Nature 447(714):719. https://doi.org/10.1038/nature05846
Kennedy SW, Jones SP (1994) Simultaneous measurement of cytochrome P4501A catalytic activity and total protein concentration with a fluorescence plate reader. Anal Biochem 222:217–223. https://doi.org/10.1006/abio.1994.1476
Kihara Y, Yustiawati TM, Gumiri S, Ardianor HT, Tanaka S, Saito T, Kurasaki M (2012) Mechanism of the toxicity induced by natural humic acid on human vascular endothelial cells. Environ Toxicol 29:916–925. https://doi.org/10.1002/tox.21819
Lin CH, Chou PH, Chen PJ (2014) Two azole fungicides (carcinogenic triadimefon and non-carcinogenic myclobutanil) exhibit different hepatic cytochrome P450 activities in medaka fish. J Hazard Mater 277:150–158. https://doi.org/10.1016/j.jhazmat.2014.05.083
Linnik PN, Ivanechko YS, Linnik RP, Zhezherya VA (2013) Humic substances in surface waters of the Ukraine. Russ J Gen Chem 83:2715–2730. https://doi.org/10.1134/S1070363213130185
Loboda A, Damulewicz M, Pyza E, Jozkowicz A, Dulak J (2016) Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell Mol Life Sci 73:3221–3247. https://doi.org/10.1007/s00018-016-2223-0
Lorenzen AKSW, Kennedy SW (1993) A fluorescence-based protein assay for use with a microplate reader. Anal Biochem 214:346–348. https://doi.org/10.1006/abio.1993.1504
Lubos E, Loscalzo J, Handy DE (2011) Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Sign 15:1957–1997. https://doi.org/10.1089/ars.2010.3586
Lushchak VI (2016) Contaminant-induced oxidative stress in fish: a mechanistic approach. Fish Physiol Biochem 42:711–747. https://doi.org/10.1007/s10695-015-0171-5
Ma Q (2011) Overview of AHR functional domains and the classical AHR signalling pathway: induction of drug metabolising enzymes. In: Pohjanvirta R (ed) The AH Receptor in Biology and Toxicology. John Wiley & Sons, pp 33–45. https://doi.org/10.1002/9781118140574.ch2
MacCarthy P (2001) The principles of humic substances: an introduction to the first principle. In: Ghabbour EA, Davies G (eds) Humic Substances: Structures, Models and Functions. The Royal Society of Chemistry, Cambridge, pp 19–30
Matsuo AY, Woodin BR, Reddy CM, Val AL, Stegeman JJ (2006) Humic substances and crude oil induce cytochrome P450 1A expression in the Amazonian fish species Colossoma macropomum (Tambaqui). Environ Sci Technol 40:2851–2858. https://doi.org/10.1021/es052437i
Meech R, Hu D-G, Miners JO, Mackenzie PI (2018) UDP-glycosyltransferases. In: McQueen CA (ed) Comprehensive Toxicology, 2nd edn. Elsevier, pp 413–434. https://doi.org/10.1016/B978-0-08-046884-6.00420-6
Nozik-Grayck E, Suliman HB, Piantadosi CA (2005) Extracellular superoxide dismutase. Int J Biochem Cell B 37:2466–2471. https://doi.org/10.1016/j.biocel.2005.06.012
OECD (2019) Test No. 203: fish, acute toxicity test. In: OECD Guidelines for the Testing of Chemicals, Section 2. OECD Publishing, Paris. https://doi.org/10.1787/9789264069961-en
Oleksiak MF, Wu S, Parker C, Qu W, Cox R, Zeldin DC, Stegeman JJ (2003) Identification and regulation of a new vertebrate cytochrome P450 subfamily, the CYP2Ps, and functional characterisation of CYP2P3, a conserved arachidonic acid epoxygenase/19-hydroxylase. Arch Biochem Biophys 411:223–234. https://doi.org/10.1016/S0003-9861(02)00734-8
Olk DC, Bloom PR, Perdue EM, McKnight DM, Chen Y, Farenhorst A, Senesi N, Chin Y-P, Schmitt-Kopplin P, Hertkorn N, Harir M (2019) Environmental and agricultural relevance of humic fractions extracted by alkali from soils and natural waters. J Environ Qual 48:217–232. https://doi.org/10.2134/jeq2019.02.0041
Orans J, Teotico DG, Redinbo MR (2005) The nuclear xenobiotic receptor pregnane X receptor: recent insights and new challenges. Mol Endocrinol 19:2891–2900. https://doi.org/10.1210/me.2005-0156
Pavek P (2016) Pregnane X Receptor (PXR)-mediated gene repression and cross-talk of PXR with other nuclear receptors via coactivator interactions. Front Pharmacol 7:456. https://doi.org/10.3389/fphar.2016.00456
Pflugmacher S, Tidwell LF, Steinberg CE (2001) Dissolved humic substances can directly affect freshwater organisms. Acta Hydroch Hydrob 29:34–40. https://doi.org/10.1002/1521-401X()29:1%3c34::AID-AHEH34%3e3.0.CO;2-8
Regoli F, Giuliani ME, Benedetti M, Arukwe A (2011) Molecular and biochemical biomarkers in environmental monitoring: a comparison of biotransformation and antioxidant defense systems in multiple tissues. Aquat Toxicol 105:56–66. https://doi.org/10.1016/j.aquatox.2011.06.014
Rhee SG (2016) Overview on peroxiredoxin. Mol Cells 39:1–5. https://doi.org/10.14348/molcells.2016.2368
Rodríguez FJ, Núñez LA (2011) Characterization of aquatic humic substances. Water Environ J 25:163–170. https://doi.org/10.1111/j.1747-6593.2009.00205.x
Saad M, Cavanaugh K, Verbueken E, Pype C, Casteleyn C, Van Ginneken C, Van Cruchten S (2016) Xenobiotic metabolism in the zebrafish: a review of the spatiotemporal distribution, modulation and activity of Cytochrome P450 families 1 to 3. J Toxicol Sci 41:1–11. https://doi.org/10.2131/jts.41.1
Schlenk D, Celander M, Gallagher EP, George S, James M, Kullman SW, van den Hurk P, Willett K (2008) Biotransformation in fishes. In: Di Giulio RT, Hinton DE (eds) The toxicology of fishes, 1st edn. CRC Press, Boca Raton, pp 153–234. https://doi.org/10.1201/9780203647295
Shamrikova EV, Vanchikova EV, Sytar TS, Zueva OM (2012) Carbon determination in natural waters and aqueous extracts from soils. Water: Chemistry and Ecology 4:88–92 [in Russian]
Steinberg CE, Paul A, Pflugmacher S, Meinelt T, Klöcking R, Wiegand C (2003) Pure humic substances have the potential to act as xenobiotic chemicals - a review. Fresen Environ Bull 12:391–401
Steinberg CE, Kamara S, Prokhotskaya VY, Manusadžianas L, Karasyova TA, Timofeyev MA, Jie Z, Paul A, Meinelt T, Farjalla VF, Matsuo AYO, Burnison BK, Menzel R (2006) Dissolved humic substances – ecological driving forces from the individual to the ecosystem level? Freshwater Biol 51:1189–1210. https://doi.org/10.1111/j.1365-2427.2006.01571.x
Steinberg CEW, Timofeyev MA, Menzel R (2009) Dissolved humic substances: interactions with organisms. In: Likens GE (ed) Encyclopedia of Inland Waters. Academic Press, pp 747–753. https://doi.org/10.1016/B978-012370626-3.00116-2
Thurman EM (1985) Amount of organic carbon in natural waters. In: Thurman EM (ed) Organic Geochemistry of Natural Water. Springer, Dordrecht, pp. 7–65. https://doi.org/10.1007/978-94-009-5095-5_2
Tonelli C, Chio IIC, Tuveson DA (2018) Transcriptional regulation by Nrf2. Antioxid Redox Sign 29:1727–1745. https://doi.org/10.1089/ars.2017.7342
Tseng H-P, Hseu T-H, Buhler DR, Wang W-D, Hu C-H (2005) Constitutive and xenobiotics-induced expression of a novel CYP3A gene from zebrafish larva. Toxicol Appl Pharm 205:247–258. https://doi.org/10.1016/j.taap.2004.10.019
Udenfriend S, Stein S, Boehlen P, Dairman W, Leimgruber W, Weigele M (1972) Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science 178:871–872. https://doi.org/10.1126/science.178.4063.871
UniProt (2021a) Proteomes - Oryzias latipes (Japanese rice fish) (Japanese killifish). https://www.uniprot.org/proteomes/UP000001038. Accessed 25 April 2021
UniProt (2021b) UniProtKB - A0A3B3H424 (A0A3B3H424_ORYLA). Protein: Peroxiredoxin-5. https://www.uniprot.org/uniprot/A0A3B3H424. Accessed 26 April 2021
UniProt (2021c) UniProtKB - A0A3B3HL19 (A0A3B3HL19_ORYLA). Protein: CYPIA1. https://www.uniprot.org/uniprot/A0A3B3HL19. Accessed 11 November 2021
UniProt (2021d) UniProtKB - A0A3B3HWP6 (A0A3B3HWP6_ORYLA). Protein: Superoxide dismutase [Cu-Zn]. https://www.uniprot.org/uniprot/A0A3B3HWP6. Accessed 26 April 2021
UniProt (2021e) UniProtKB - A5JL98 (A5JL98_ORYLA). Protein: Submitted name: Cytochrome P450 2P3. https://www.uniprot.org/uniprot/A5JL98. Accessed 5 November 2021
UniProt (2021f) UniProtKB - H2L5D9 (H2L5D9_ORYLA). Protein: Glutathione peroxidase. https://www.uniprot.org/uniprot/H2L5D9. Accessed 26 April 2021
UniProt (2021g) UniProtKB - H2M443 (H2M443_ORYLA). Protein: Peroxiredoxin-1. https://www.uniprot.org/uniprot/H2M443. Accessed 26 April 2021
UniProt (2021h) UniProtKB - H2MDG9 (H2MDG9_ORYLA). Protein: Superoxide dismutase. https://www.uniprot.org/uniprot/H2MDG9. Accessed 26 April 2021
UniProt (2021i) UniProtKB - H2MHT9 (H2MHT9_ORYLA). Protein: Peroxiredoxin-1. https://www.uniprot.org/uniprot/H2MHT9. Accessed 26 April 2021
UniProt (2021j) UniProtKB - H2MZ57 (H2MZ57_ORYLA). Protein: Submitted name: Cytochrome P450 3A40. https://www.uniprot.org/uniprot/H2MZ57 Accessed 11 November 2021
Valencia S, Marín JM, Restrepo G, Frimmel FH (2013) Application of excitation–emission fluorescence matrices and UV/Vis absorption to monitoring the photocatalytic degradation of commercial humic acid. Sci Total Environ 442:207–214. https://doi.org/10.1016/j.scitotenv.2012.10.058
Whyte JJ, Jung RE, Schmitt CJ, Tillitt DE (2000) Ethoxyresorufin-O-deethylase (EROD) activity in fish as a biomarker of chemical exposure. Crit Rev Toxicol 30:347–570. https://doi.org/10.1080/10408440091159239
Wiegand C, Pehkonen S, Akkanen J, Penttinen OP, Kukkonen JV (2007) Bioaccumulation of paraquat by Lumbriculus variegatus in the presence of dissolved natural organic matter and impact on energy costs, biotransformation and antioxidative enzymes. Chemosphere 66:558–566. https://doi.org/10.1016/j.chemosphere.2006.05.048
Younus H (2018) Therapeutic potentials of superoxide dismutase. Int J Health Sci 12:88–93
Zhang L, Dong X, Wang C, Zuo Z, Chen M (2017) Bioaccumulation and the expression of hepatic cytochrome P450 genes in marine medaka (Oryzias melastigma) exposed to difenoconazole. J Environ Sci 52:98–104. https://doi.org/10.1016/j.jes.2016.03.011
Zhao Q, Shi F, Zhu L (2017) Prometryn and humic acid induce Cytochrome P450 1A expression in Danio rerio (zebrafish). Ecotox Environ Safe 135:40–47. https://doi.org/10.1016/j.ecoenv.2016.09.011
Acknowledgements
We are grateful to Donald Tillitt (USGS CERC) for encouraging this study, Roman Fedorov and Dmitriy Philippov (IBIW RAS) for their help with the collection of liver samples, and Lyudmila Bakina (SPC RAS) for the analysis of organic carbon in water samples. We thank Olga Tikhonova for protein profiling data curation, Elena Hryapova for liver sample preparation, and Victor Zgoda for HPLC-MS/MS analysis of the samples. HPLC-MS/MS analysis was performed at the “Human Proteome” Core Facility (IBMC, Moscow, Russia).
Funding
This research was funded by the Russian Science Foundation, grant number 20–76-00030.
Author information
Authors and Affiliations
Contributions
Conceptualisation, Victoria Yurchenko; investigation, Victoria Yurchenko and Alexey Morozov; writing – original draft preparation, Victoria Yurchenko and Alexey Morozov; writing – review and editing, Victoria Yurchenko and Alexey Morozov.
Corresponding author
Ethics declarations
Ethics approval
Fish were treated according to the procedure for the use of fishes in research of the Laboratory of Physiology and Toxicology of Aquatic Animals and approved by the Ethics Committee of Papanin Institute for Biology of Inland Waters Russian Academy of Sciences.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yurchenko, V., Morozov, A. Responses of hepatic biotransformation and antioxidant enzymes in Japanese medaka (Oryzias latipes) exposed to humic acid. Fish Physiol Biochem 48, 1–13 (2022). https://doi.org/10.1007/s10695-021-01034-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-021-01034-4