Abstract
Background BRCA1 recurrent mutations have rarely been assessed in non-founder populations. Still, identifying such mutations could be important for designing genetic testing strategies for high-risk breast/ovarian cancer families in non-founder populations. Objective To determine whether the recurrent BRCA1 Y101X mutation identified in Yoruban breast cancer patients represents a single historical mutation event, and determine the prevalence of this mutation in a hospital based cohort. Methods 365 breast cancer patients and 177 controls of Yoruban ancestry from Nigeria, unselected for age of onset or family history were screened for the BRCA1 Y101X mutation. The haplotypes on which the Y101X mutation occurred were characterized using microsatellite markers and single-nucleotide polymorphisms (SNPs). Phase ambiguity was resolved using allele-specific PCR. Results The BRCA1 Y101X mutation was detected in four Yoruban patients with no documented family history of breast cancer among a cohort of 365 (1.1, 95% C.I. = 0.43–2.78%) unrelated Yoruban breast cancer patients. This study reveals the four Y101X mutations occur on a single, rare haplotype. Further characterization in a patient of European ancestry with a strong family history of breast/ovarian cancer revealed the same Y101X mutation on the same haplotype as those in the Yoruban carriers. These observations suggest the Y101X mutations identified in the Yoruban patients may have originated from a single mutation event. Conclusions BRCA1 Y101X is the first reported recurrent mutation occurring in patients of African ancestry for which prevalence has been determined. Identification of this mutation in a woman of European ancestry with strong family history of breast/ovarian suggests further that this mutation occurred once, probably many generations ago.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
About 5–10% of breast cancer cases are associated with inherited susceptibility, and only a subset of these are due to germline mutations in the BRCA1 and BRCA2 tumor suppressor genes [1]. These mutations are often highly penetrant and are among the strongest genetic predictors of breast cancer occurrence [2–4]. Recurrent BRCA1 and BRCA2 germline mutations have been described primarily in populations with specific ancestries, reflecting founder effects, in which single mutations in small ancestral populations with limited genetic diversity may become frequent alleles in descendant populations. The identification of founder mutations is important for designing genetic testing strategies for mutations in genes associated with inherited elevated disease risk, like BRCA1. For example, the Ashkenazi Jewish founder mutations BRCA1 185delAG, 5382insC and BRCA2 6174delT, constitute an “Ashkenazi panel” used to expedite BRCA1 and BRCA2 mutation testing in Ashkenazi Jewish families at high risk of breast and ovarian cancer [4–6]. BRCA1 and BRCA2 founder mutations have been identified in different European populations, including Belgian [7], French-Canadian [8], German [9], Spanish [10] and Swedish [11] populations. Several studies have also reported BRCA1 and BRCA2 founder mutations in Asian populations including Chinese [12], Japanese [13], Malay [14], Thai [15] and Filipino [16] populations. However, thus far, only one mutation (BRCA1 943ins10) has been identified as a potential founder mutation in patients of West African ancestry [17], and the frequency of this mutation in breast cancer patients of African ancestry has not been determined. For patients of African ancestry to benefit from population-specific risk assessment that has been so valuable in other populations, it is important to identify and characterize recurrent mutations that may contribute to cancer risk in women of African ancestry.
Here we describe a recurrent mutation in Nigerian breast cancer patients of Yoruban ancestry, BRCA1 Y101X (422T>G), that likely arose from a single mutation event in the Yoruba population in Nigeria. The mutation was further characterized in a patient of European ancestry with a strong family history of early-onset breast and ovarian cancer. This represents the first report of a recurrent, potentially ancient mutation among indigenous Yoruban peoples which has implications for the design of genetic testing strategies in such a low resource country.
Materials and methods
Patients
The study setting and design have been described in detail elsewhere [18]. Briefly, breast cancer cases were identified through the Surgical Oncology and Radiotherapy units of the University College Hospital (UCH), Ibadan, Nigeria. All consecutive female breast cancer cases aged 18 and above, with a clinical diagnosis of invasive breast cancers between March 1998 and July 2006, were eligible. The majority of eligible patients approached agreed to participate in the study, with a refusal rate of 4%. During the period of case enrollment, a community adjoining the hospital was randomly selected by ballot from the list of all the communities in the hypothetical catchment area of the hospital. This community is stable, socio-economically diverse and represents the diversity of patients seen at the UCH. Inclusion criteria for the controls were female, aged 18 years or older, absence of any type of cancer at the time of recruitment, and ability to give informed consent [18].
Genomic DNA was prepared from peripheral blood from breast cancer patients and control subjects using Puregene™ DNA isolation kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions [1].
Mutation identification and mutation genotyping
The BRCA1 Y101X (422T>G), exon 7) mutation was identified as part of a characterization of BRCA1 genomic variants in the Nigerian population [1 and unpublished observations]. Patients were screened for heterozygosity in BRCA1 by denaturing high-performance liquid chromatography (DHPLC), using standard polymerase chain reaction (PCR) and a Transgenomic WAVE® 3500HT Nucleic Acid Fragment Analysis System (Transgenomic, Inc., Omaha, NE, USA). PCR fragments resulting in heteroduplex molecules identified by DHPLC were sequenced directly using a fluorescent dye dideoxy chain terminator (BigDye 3.1) reaction and products were analyzed on an Applied Biosystems 3730XL 96-capillary DNA sequencer (both from Applied Biosystems, Foster City, CA, USA).
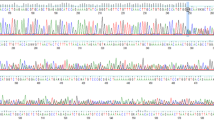
After two BRCA1 Y101X mutations were detected using DHPLC pre-screening, BRCA1 exon 7 PCR amplicons were sequenced directly in 365 breast cancer patients, including the patients previously screened by DHPLC (Fig. 1). 177 unaffected Yoruban control subjects were also analyzed by direct sequencing of BRCA1 exon 7.
Microsatellite markers and SNPs genotyping
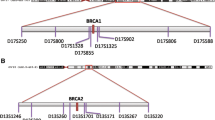
Genotype analysis was performed using microsatellite markers within or near BRCA1 (tel-D17S1329-D17S1325-D17S1326-D17S1327-D17S1323-D17S1322-D17S855-D17S1321-D17S846-D17S800-D17S250-Cen) (Fig. 2) [19]. PCR products representing these markers were labeled with fluorescent dye and mixed with fluorescently labeled size markers and run on an Applied Biosystems 3730XL 96-capillary DNA sequencer. Results were analyzed using Genemapper 3.7 (Applied Biosystems).
Genotype analysis was also performed using previously described BRCA1 tag SNPs and missense SNPs: tel-rs4793211-rs1800062-rs799914-rs799923-rs799918-rs1799950-rs799917-rs2227945-rs16942-rs8176166-rs3737559-rs2236762-rs1799966-rs8176267-Cen [20] (Fig. 2). The haplotypes were constructed using PHASE 2.1 software [21, 22], which uses a Bayesian algorithm to predict the haplotype based on genotype data.
Allele-specific PCR
The 2.2 kb genomic DNA fragment containing the D17S1323 allele on the same chromosome as the T allele of the heterozygous rs8176166 SNP in all four BRCA1 Y101X carriers was amplified using the allele-specific primer 5′-TCAGTTTGGGATGATGAAAA-3′ and the reverse primer, 5′-TTGAAGCAACTTTGCAATGAG-3′. The allele-specific PCR was performed using the following program: one cycle of denaturation at 95°C for 10 min, 35 cycles of denaturation at 95°C for 40 s, annealing at 57°C for 40 s, extension at 72°C for 2 min and a final extension step of 72°C for 5 min. The reaction was performed in a volume of 20 μl, at a final concentration of 0.2 mM MgCl2, using AmpliTaq Gold manufactured by Roche (Applied Biosystems, Foster City, California, USA). The PCR product was diluted 1:10,000 and used as the template for a nested amplification of a small DNA segment containing D17S1323. The primers for this second amplification are as follows: 5′/56-FAM/TAGGAGATGGATTATTGGTG-3′ and 5′-AAGCAACTTTGCAATGAGTG-3′. The second PCR amplification was performed using the following program: one cycle of denaturation at 95°C for 10 min, 28 cycles of denaturation at 95°C for 40 s, annealing at 57°C for 40 s, an extension step of 72°C for 30 s; and a final extension step of 72°C for 1 min. Microsatellite marker allele calling was performed as described above.
Results
BRCA1 Y101X (422T>G) mutations were detected in a total of four out of 365 Yoruban breast cancer cases unselected for age of onset or family history (1.1, 95% C.I. = 0.43–2.78%) (Fig. 1a). The mean age of diagnosis of the Yoruban breast cancer cases is 47.8 years and the ages of diagnosis range from 16 to 78 year. All four detected BRCA1 Y101X mutations were confirmed by restriction enzyme digestion (Data not shown). All four patients carrying the mutation were self-reported as Yoruban, with no apparent family history of breast cancer. Ages of diagnosis in the Y101X carriers were 32 years (one patient), 50 years (two patients) and 71 years (one patient). None of the 177 unaffected Yoruban control individuals carried the BRCA1 Y101X mutation.
To determine whether all four occurrences of the BRCA1 Y101X mutation descended from a single ancestral mutation event, we genotyped and analyzed the following microsatellite markers: tel-D17S1329-D17S1325-D17S1326-D17S1327-D17S1323-D17S1322-D17S855-D17S1321-D17S846-D17S800-D17S250-cen, spanning a 4.6 Mb interval containing BRCA1 (Table 1, Fig. 2). The UniSTS database lists D17s1326 and D17s1327 as each mapping to two locations by e-PCR. The positions shown in Fig. 2 are consistent with entries in the Celera database, build 36.3 (http://www.ncbi.nlm.nih.gov). The positions D17s1326 and D17s1327 are mapped to chromosome 17, position: 38041918-38042018 (bp) and position: 38032314-38032443 (bp) in the Celera database, respectively. Family members of BRCA1 Y101X mutation carriers were unavailable for genetic analysis, so haplotypes could not be determined directly from markers in relatives. For D17S1326-D17S1327-D17S1323-D17S1322-D17S855-D17S1321, which extends over a 0.53–1.74 Mb region, all the mutation carriers have a shared allele for each marker except at D17S1323, where three of four mutation carriers share either allele 5 or allele 7 of this marker. This exception is most likely due to a microsatellite mutation event after the initial BRCA1 Y101X mutation since the downstream markers in these four carriers have common alleles. Thus, the haplotype of a single ancestral chromosome may be inferred for the BRCA1 Y101X mutation as illustrated in Table 1. The genotypes of markers tel-D17S1326-D17S1327-D17S1323-D17S1322-D17S855-cen were also analyzed in 79 control subjects. The control frequencies of the putative BRCA1 Y101-linked alleles for each marker are: D17S1326 allele 3, 25/154 (16.1%); D17S1327 allele 12, 5/152 (3.3%); D17S1322, allele 4, 46/134 (34.3%) and allele 3, 41/134 (30.6%); D17S855 allele 3, 10/150 (6.7%). Note that most of the alleles shared by the four BRCA1 Y101X carriers at these polymorphic markers are infrequent in the control chromosomes. Additionally, none of the 79 control subjects had genotypes that included all the alleles of the consensus haplotypes. Thus, it is unlikely that the BRCA1 Y101X mutation arose independently on several chromosomes.
BRCA1 Y101X resides on a single high-resolution SNP haplotype
To further describe the ancestral chromosome on which the BRCA1 Y101X mutation occurred, we constructed SNP-based haplotypes for the four BRCA1 Y101X carriers and wild type controls using nine BRCA1 tag SNPs and five missense SNPs as described previously [20] and PHASE 2.1 software (Table 2a). All the Y101X carriers and control females are homozygous at five of these SNPs: rs1800062, rs799923, rs1799950, rs2227945, and rs8176267. Using the HapMap Genome Browser (B36) (www.hapmap.org) we found that rs799923 and rs1799950 have common minor alleles in the CEPH (Utah residents with ancestry from northern and western Europe) population and are informative. Therefore, haplotype analysis includes these two SNPs, but does not include the data at rs1800062, rs2227945, and rs8176267. Every patient harboring BRCA1 Y101X was predicted with a probability over 93.8% [21, 22] to have a common mutation haplotype: C-G-G-G-T-A-C-C-C-T-C at tel-rs799914-Y101X-rs799923-rs799918-rs1799950-rs799917-rs16942-rs8176166-rs3737559-rs2236762-rs1799966-cen (Table 2b). Twenty-one out of 354 control chromosomes (5.9, 95% C.I. = 3.7–8.4%) were predicted to carry the same haplotype without the mutation C-T-G-G-T-A-C-C-C-T-C. When rs4793211 was included in the analysis, the PHASE program still predicted that all four mutations were most likely on a single mutation haplotype T-C-G-G-G-T-A-C-C-C-T-C, but with a lowered probability. This extended haplotype is present on 16 control chromosomes has a frequency of 4.5% in control chromosomes (4.5, 95% C.I. = 2.8–7.2%). The relative rarity of this haplotype supports our previous observation using microsatellite markers, that the four BRCA1 Y101X mutations probably arose from a single mutation event on a fairly rare chromosome.
Molecular determination of Y101X-D17S1323 allele linkage
Because two D17S1323 alleles (5 and 7) are both potentially linked to the BRCA1 Y101X mutation, we performed allele-specific PCR in the four mutation carriers to amplify D17S1323-containing fragments from the T allele of rs8176166. Assuming haplotype 10 (Table 2a) correctly describes the markers flanking the BRCA1 Y101X mutation, we would predict that the T allele of rs8176166 is linked to the wild type T allele at codon 101. The results indicated that the allele T at rs8176166 is on the same chromosome as the non-7 allele at D17S1323 in the mutation carriers (Fig. 3). This suggests that the allele C at rs8176166 is on the same chromosome as D17S1323 allele 7 in three carriers. In patient 655318 the allele C is linked to D17S1323 allele 5, suggesting a mutation event at D17S1323 occurred in carrier 655318 after the initial single Y101X mutation event. As the C allele of rs8176166 is in phase with the mutant G allele in the BRCA1 Y101X codon, we conclude that the founding mutation occurred on a chromosome containing the D17S1323 allele 7. Similarly, either allele three or allele four of D17S1322 could be linked to the BRCA1 Y101X mutation. However, this marker is not accessible by the long-range PCR strategy employed for D17S1323, so the allele that lies within the ancestral BRCA1 Y101X haplotype is as yet unresolved.
Allele-specific PCR identifies the D17S1323 allele linked to BRCA1 Y101X. A chromatogram of D17S1323 in BRCA1 Y101X mutation carrier 925728 (a), and the same sample amplified with the rs8176166 T-specific allele PCR primer (b). While allele-specific PCR introduced variable background peak intensities, it is clear that D17S1323 allele 7 is not amplified by the rs8176166 T allele-specific PCR
Characterization of BRCA1 Y101X in a female of European ancestry
To further characterize the BRCA1 Y101X mutation, we identified another carrier of the mutation through Myriad Genetics, a 46-year-old patient with a strong family history of early-onset breast and ovarian cancer. The patient is of European ancestry, including Irish, English, German, and Yugoslavian background as illustrated in her pedigree (Fig. 4). All of the studied SNPs surrounding the BRCA1 Y101X were found to be homozygous for this carrier of European ancestry (Table 2b). The genotypes of the SNPs suggest that BRCA1 Y101X in this carrier occurs on the same haplotype as the BRCA1 Y101X mutations in the four Yoruban carriers (Table 2b). From the allele and genotype frequencies of the SNPs, it is unlikely that the Y101X in this patient of European ancestry occurred independently from the lineage of the mutations in the Yoruban patients. Thus, our data suggests the BRCA1 Y101X mutations of European and Yoruban ancestry are related and possibly arose from a single mutation event in history (Table 2c). However, the genotypes of the microsatellite markers indicates that the four Yoruban BRCA1 Y101X carriers do not share a common allele with the patient of European ancestry at the surrounding microsatellite markers: D17S1327, D17S1323, D17S855, and D17S1321 (Table 1). Of note, microsatellite marker D17S1323, which falls within the conserved region of the ancestral SNP haplotype, shares no alleles in common with the putative ancestral microsatellite haplotype. It is possible that the alleles linked to BRCA1 Y101X were altered in the carrier of European ancestry after the common ancestral mutation event, due to the higher mutation rates of microsatellite markers.
Discussion
The founder mutations BRCA1 185delAG and 5382insC are used for efficient genetic screening strategies in Ashkenazi Jews at high risk of breast cancer. BRCA1 founder mutations have been reported in many populations such as Belgian, Dutch, German, Swedish, Japanese, Malay and Chinese. In populations of African ancestry, only the BRCA1 943ins10 has been described as a recurrent, potential founder mutation from western Africa. Still, the population frequency of this mutation has not been determined [23, 24]. In the present study, we found a recurrent BRCA1 mutation in four out of 365 unrelated Yorubanbreast cancer patients (1.1, 95% C.I. = 0.43–2.78%). This mutation has a frequency in this hospital-based Yoruban breast cancer patient population comparable to the founder mutation 1135insA in Norwegian ovarian cancer cases unselected for age or family history (0.81, 95% C.I. = 0.35–1.89%) [25]. The haplotype of the flanking SNPs and the common alleles at microsatellite markers spanning over a 0.53–1.74 Mb region, suggest that the BRCA1 Y101X mutation of Yoruban ancestry is unlikely to have arisen multiple times as independent events, but rather represents a single mutation in an ancestor common to all four Yoruban patients examined. This suggestion is supported by the observation that the single haplotype on which the BRCA1 Y101X mutation occurs is found in only 4.5% of the control chromosomes, indicating a low probability of multiple BRCA1 Y101X mutations occurring independently on a frequently occurring haplotype. The genotypes and haplotypes of the flanking SNPs suggest BRCA1 Y101X in the patient of European ancestry examined likely arose from the same ancestral mutation that also gave rise to the BRCA1 Y101X in the Yoruban patients examined.
When recurrent mutations are suspected of representing founder mutations, it is useful to estimate the number of generations since the single mutation event [16, 26]. Unfortunately, the available data do not allow an age estimation for the mutation.
Microsatellite markers have much higher mutation rates than SNPs, and most microsatellite markers are located farther from BRCA1 Y101X than the most useful SNPs (the exceptions being D17S1323 and D17S1322). Thus, much more genetic divergence is expected to have occurred at the microsatellite markers in the history of this mutation. This can explain why the carrier of European ancestry and Yoruban carriers do not share the common alleles at several surrounding microsatellite markers, but the BRCA1 Y101X mutations in both the carrier of European ancestry and the Yoruban carriers are on the same SNPs haplotype. The genotypes of the SNPs and the microsatellite markers for the BRCA1 Y101X carrier of European ancestry further suggest that BRCA1 Y101X might be an ancient mutation.
BRCA1 Y101X has been reported twice in the Open Access On-Line Breast Cancer Mutation Database as a deleterious mutation. The ethnic background of these reported case are unknown. By convention, BRCA1 Y101X is described as a nonsense mutation because of the predicted effect of the T>G change on codon 101. However, the 422T>G base change resulting in the Y101X codon change occurs on the second base of exon 7, (Fig. 1) which could potentially alter the mRNA splicing pattern. The molecular basis of the Y101X gene defect may therefore be more complex than a single predicted protein truncation. Unfortunately, mRNA samples for molecular characterization are unavailable from these patients. However, the family history of early-onset breast and ovarian cancer associated with the European BRCA1 Y101X carrier suggests this mutation may be a functional null mutation.
Founder mutations are typically an indicator of low genetic diversity, often associated with bottlenecks in population size. However, the Yoruba population studied here exhibits a greater genetic diversity of BRCA1-associated markers than most other populations studied (Table 2) [20], and the Yoruba population is large and ancient. It is therefore possible that the four mutation carriers reflect a population substructure that may not represent potential Nigeria-wide mutation frequency. In particular, because the BRCA1 Y101X mutation carriers were identified in the same hospital setting, it is possible that that these individuals were more closely related than could have been anticipated from questionnaire responses. Future work will further examine the population frequency of this mutation.
In conclusion, the identification of this recurring BRCA1 mutation in Yoruban breast cancer patients underscores the need for assessment of recurrent mutations in non-founder populations that could potentially be found in other populations.
References
Fackenthal JD, Sveen L, Gao Q et al (2005) Complete allelic analysis of BRCA1 and BRCA2 variants in young Nigerian breast cancer patients. J Med Genet 42(3):276–281. doi:10.1136/jmg.2004.020446
Warner E, Foulkes W, Goodwin P et al (1999) Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst 91(14):1241–1247. doi:10.1093/jnci/91.14.1241
Ford D, Easton DF, Stratton M et al (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The breast cancer linkage consortium. Am J Hum Genet 62(3):676–689. doi:10.1086/301749
King MC, Marks JH, Mandell JB et al (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302(5645):643–646. doi:10.1126/science.1088759
Friedman LS, Szabo CI, Ostermeyer EA et al (1995) Novel inherited mutations and variable expressivity of BRCA1 alleles, including the founder mutation 185delAG in Ashkenazi Jewish families. Am J Hum Genet 57(6):1284–1297
Neuhausen S, Gilewski T, Norton L et al (1996) Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet 13(1):126–128. doi:10.1038/ng0596-126
Claes K, Machackova E, De Vos M et al (1999) Mutation analysis of the BRCA1 and BRCA2 genes in the Belgian patient population and identification of a Belgian founder mutation BRCA1 IVS5+3A>G. Dis Markers 15(1–3):69–73
Tonin PN, Mes-Masson AM, Futreal PA et al (1998) Founder BRCA1 and BRCA2 mutations in French Canadian breast and ovarian cancer families. Am J Hum Genet 63(5):1341–1351. doi:10.1086/302099
Hartmann C, John AL, Klaes R et al (2004) Large BRCA1 gene deletions are found in 3% of German high-risk breast cancer families. Hum Mutat 24(6):534. doi:10.1002/humu.9291
Vega A, Campos B, Bressac-De-Paillerets B et al (2001) The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum Mutat 17(6):520–521. doi:10.1002/humu.1136
Bergman A, Einbeigi Z, Olofsson U et al (2001) The western Swedish BRCA1 founder mutation 3171ins5; a 37 cM conserved haplotype of today is a reminiscence of a 1500-year-old mutation. Eur J Hum Genet 9(10):787–793. doi:10.1038/sj.ejhg.5200704
Khoo US, Chan KY, Cheung AN et al (2002) Recurrent BRCA1 and BRCA2 germline mutations in ovarian cancer: a founder mutation of BRCA1 identified in the Chinese population. Hum Mutat 19(3):307–308. doi:10.1002/humu.9015
Sekine M, Nagata H, Tsuji S et al (2001) Mutational analysis of BRCA1 and BRCA2 and clinicopathologic analysis of ovarian cancer in 82 ovarian cancer families: two common founder mutations of BRCA1 in Japanese population. Clin Cancer Res 7(10):3144–3150
Lee AS, Ho GH, Oh PC et al (2003) Founder mutation in the BRCA1 gene in Malay breast cancer patients from Singapore. Hum Mutat 22(2):178. doi:10.1002/humu.9162
Patmasiriwat P, Bhothisuwan K, Sinilnikova OM et al (2002) Analysis of breast cancer susceptibility genes BRCA1 and BRCA2 in Thai familial and isolated early-onset breast and ovarian cancer. Hum Mutat 20(3):230. doi:10.1002/humu.9049
De Leon Matsuda ML, Liede A, Kwan E et al (2002) BRCA1 and BRCA2 mutations among breast cancer patients from the Philippines. Int J Cancer 98(4):596–603. doi:10.1002/ijc.10194
Mefford HC, Baumbach L, Panguluri RC et al (1999) Evidence for a BRCA1 founder mutation in families of West African ancestry. Am J Hum Genet 65(2):575–578. doi:10.1086/302511 letter
Adebamowo CA, Ogundiran TO, Adenipekun AA et al (2003) Obesity and height in urban Nigerian women with breast cancer. Ann Epidemiol 13(6):455–461. doi:10.1016/S1047-2797(02)00426-X
Neuhausen SL, Mazoyer S, Friedman L et al (1996) Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am J Hum Genet 58(2):271–280
Freedman ML, Penney KL, Stram DO et al (2005) A haplotype-based case-control study of BRCA1 and sporadic breast cancer risk. Cancer Res 65(16):7516–7522. doi:10.1158/0008-5472.CAN-05-0132
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68(4):978–989. doi:10.1086/319501
Stephens M, Donnelly P (2003) A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73(5):1162–1169. doi:10.1086/379378
Olopade OI, Fackenthal JD, Dunston G et al (2003) Breast cancer genetics in African Americans. Cancer 97(Suppl 1):236–245. doi:10.1002/cncr.11019
Panguluri RC, Brody LC, Modali R, Utley K, Adams-Campbell L, Day AA et al (1999) BRCA1 mutations in African Americans. Hum Genet 105(1–2):28–31. doi:10.1007/s004390051059
Dorum A, Hovig E, Trope C, Inganas M, Moller P et al (1999) Three percent of Norwegian ovarian cancers are caused by BRCA1 1675delA or 1135insA. Eur J Cancer 35(5):779–781
Couch FJ, Farid LM, DeShano ML et al (1996) BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet 13(1):123–125. doi:10.1038/ng0596-123
Acknowledgements
This work was supported by grants from R01 CA 89085-01A1 the National Cancer Institute, the Falk Medical Research Trust, and the University of Chicago Cancer Research Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
B. Zhang, J. D. Fackenthal contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, B., Fackenthal, J.D., Niu, Q. et al. Evidence for an ancient BRCA1 mutation in breast cancer patients of yoruban ancestry. Familial Cancer 8, 15–22 (2009). https://doi.org/10.1007/s10689-008-9205-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-008-9205-9