Abstract
Identifying and using heterotic loci in breeding programs is crucial for increasing crop yield. In this study, we planted 68 inbred lines and their 132 testcrosses at Wuhan and Xiangyang, China. We performed a phenotypic analysis of nine yield-related traits, including plant height, main inflorescence length, branch number, seeds per silique, effective silique number on the main inflorescence, thousand seed weight, biomass yield per plant, seed yield per plant, and harvest index (HI). Additionally, the mid-parent heterosis (MPH) and better-parent heterosis (BPH) for each trait were evaluated. The genetic distance (GD) calculated using high-throughput single nucleotide polymorphisms (SNPs) showed no correlation with HI heterosis, and the difference in GD in the two groups (H-154 and H-155) was not correlated with the difference in HI heterosis levels. The MPH and BPH of HI were significantly correlated with the MPH and BPH of biomass yield per plant, but not with the other traits. Our results indicate that yield-related traits heterosis, including HI heterosis, are extremely complex. The effectiveness of predicting heteroses with GD based on SNPs may be limited, though the SNPs can be used to instead of older markers for high-throughput and accurate genotyping. Our findings may contribute a better understanding to rapeseed breeding and the natural variations of HI heterosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heterosis refers to the superior performance of hybrids over either parent in terms of plant biomass, fertility, and other characteristics (Riddle and Birchler 2008). Fully characterizing the genetic mechanism mediating heterosis may help in the development of methods to increase plant and animal productivity. Dominance (Xiao et al. 1995), overdominance (Shull 1946), epistasis (Yu et al. 1997; Hua et al. 2003), and epigenomic and small RNA (Chen 2013) models were proposed to explain the genetic basis of heterosis in model or classical hybrid plants like Arabidopsis thaliana, maize, and rice. However, none of the current genetic models can completely explain the heterosis phenomenon, especially for polyploid plants, such as rapeseed. Calculating genetic distance (GD) based on DNA markers has typically been used to explain the performance of rapeseed hybrids. One of the common features of the aforementioned studies is the use of traditional markers, such as morphological characteristics (Ali et al. 1995), restriction fragment length polymorphisms (Diers et al. 1996; Liu et al. 2002), sequence-related amplified polymorphisms (Riaz et al. 2001), random amplified polymorphic DNAs (Yu et al. 2005; Teklewold and Becker 2006), amplified fragment length polymorphisms (Qian et al. 2007), and simple sequence repeats (Xing et al. 2014).
Rapeseed (Brassica napus L.; AACC, 2n = 38) consists of a highly complex genome structure and has an extremely high phenotypic and genetic diversity. Therefore, low-density DNA markers cannot be used to comprehensively examine the genetic diversity. Fortunately, expressed sequence tag data, array analyses, amplicon resequencing, sequenced genomes, and next-generation sequencing technologies have been used to identify high-throughput single nucleotide polymorphisms (SNPs) in a given plant (Ganal et al. 2009). This has enabled researchers to scan genomic regions with high marker densities to screen for genetic diversity (Blair et al. 2013), and perform marker-assisted selection (Yan et al. 2010) and genome-wide association studies (McCarthy et al. 2008). Recently, high-throughput SNPs covering the whole B. napus genome were identified by Emei Tongde Co. (Beijing) using the Brassica 60 K Illumina® Infinium SNP array (Snowdon and Iniguez Luy 2012). Sang et al. (2015) estimated the correlation between GD based on 40,201 SNPs and 10 yield-related traits heterosis. They determined that GD has a very significant positive correlation with heterosis for traits with high and universal heterosis, including plant yield. There is currently no information regarding harvest index (HI) heterosis and the relationship between HI heterosis and heteroses of other yield-related traits, let alone the relationship between GD based on genomic SNPs and HI heterosis.
In this study, we completed a phenotypic analysis of nine traits [plant height (PH), main inflorescence length (IL), branch number (BN), seeds per silique (SS), effective silique number on the main inflorescence (ISN), thousand seed weight (TSW), biomass yield per plant (BY), seed yield per plant (SY), and HI] and their heteroses with 68 inbred lines and 132 testcrosses at two locations. The purpose of the present study was to investigate: (1) the phenotypic variation of HI heterosis, (2) the GD between the parental lines, (3) the relationship between HI heterosis and heteroses of other yield-related traits, and (4) the relationship between GD and HI heterosis.
Materials and methods
Plant materials and field experiments
The parental lines were derived from an association mapping population containing 155 Chinese winter rapeseed lines genotyped with the 60 K Illumina® Infinium SNP array (Luo et al. 2015). Of the parental lines, 153 were crossed with the other two lines (L154 and L155) by hand crossing during flowering in the 2010/2011 growing season in Wuhan, China. We collected 218 F1 seeds from the crosses of 149 lines with L154 or L155 at maturity. The hybrids along with the other lines were grown in a randomized complete block design with two replications at Wuhan (29.58°N, 113.53°E) and Xiangyang (32.01°N, 112.08°E) along the middle reaches of the Yangtze River, China in the 2012/2013 growing season. A plot consisting of three rows was used for the two replications with 12 plants per row. We used a spacing of 30 × 20 cm between rows and between plants within a row at both locations. The mature plants from the middle of each plot were selected for trait evaluation, and the mean value of each trait was used for analyses. Measurements for PH, IL, BN, SS, ISN, TSW, BY, and SY were completed as described by Shi et al. (2011). The HI (%) was calculated as the ratio of SY to BY. To ensure accurately assess phenotypes, the heterosis values for 132 hybrids produced from 66 lines crossed with L154 and L155, a completed testcross mating population, were used to study in the paper (Online Resources 1 and 2).
Genetic distance analysis
The genetic diversity of the parental lines was estimated using 7600 SNPs with AA and BB frequencies >0.03 (i.e., monomorphic), call frequency >0.9, and minor frequency >0.05 (Luo et al. 2015). The GD between parental lines was calculated using these SNPs and the unweighted pair-group method with arithmetic mean of NTSYSpc Version 2.10e software (Rohlf 2000). The polymorphism information content of the 7600 SNPs was estimated using PowerMarker version 3.51 (Liu and Muse 2005).
Statistical analysis
The hybrids were classified into two groups (H-154 and H-155) according to different testers, and phenotypic variance in two groups (H-154 and H-155) was analyzed, respectively. The mid-parent heterosis (MPH) was calculated as: MPH (%) = (F1-MP)/MP × 100, and the better-parent heterosis (BPH) was calculated as: BPH (%) = (F1-BP)/BP × 100, where MP refers to the mean of the two parents and BP corresponds to the value of the better parent. The variances of the additive effect (A), dominant effect (D), and additive-by-additive effect (AA), and their interaction with the environment (AE, DE, and AAE) were estimated using the QGA Station Version 1.0 software (Zhu and Weir 1994). The significant difference between group mean values for each trait heterosis was estimated by t test. The differences in GD and heterosis levels were determined by the value difference between the two groups according to different testers. The correlation analysis was performed with SPSS version 19.0 (IBM Corp., Armonk, NY, USA). P values of 0.05 and 0.01 were considered to be statistically significant and highly significant, respectively.
Results
Heterosis performance of hybrids
Hybrids were classified into two groups (H-154 and H-155). For MPH, H-154 had significantly higher mean values than H-155 for SY, BY, IL, ISN, and SS (P = 0.01) and HI (P = 0.05) (Online Resource 1). For BPH, H-154 also had significantly higher mean values than H-155 for SY, BN, IL, PH, ISN, and SS (P = 0.01) and BY and HI (P = 0.05) (Online Resource 2).
The average MPH and BPH values showed a positive trend for all traits (Table 1). The MPH values ranged from −30 % for SY of H-155 to 82 % for SY of H-154, and the BPH values ranged from −33 % for SY of H-155 to 69 % for SY of H-154. The ISN trait had the highest average MPH (44 %) and BPH (33 %) among all traits. The average MPH values for HI were 14 and 12 % in H-154 and H-155, respectively, and were up to 43 % for certain testcrosses in H-155. The average BPH values for HI were 8 and 4 % in H-154 and H-155, respectively, and were up to 39 % for certain testcrosses in H-155.
Estimation of variance ratios for traits
Results of the two-factor analysis of variance for the performance of the parental lines and hybrids are provided in Online Resource 3. The residual variance ratio of yield-related traits ranged from 10 to 33 %. The A and D variance ratios were significant for all traits (P = 0.01), ranging from 26 to 49 % and 24 to 39 %, respectively. However, AA was not significant for all traits. Regarding the interactions with the environment, AE was statistically significant for all traits except HI (P = 0.01) and there was a small but significant DE value for TSW (4 %) and BY (6 %) (P = 0.01), but not for the other traits. The AAE values were only significant for BN and HI (P = 0.01), with HI (8 %) having a much higher AAE than BN (1 %).
Correlations between HI heterosis and heteroses of other yield-related traits
In general, the MPH and BPH of HI were significantly correlated with the MPH and BPH of BY (P = 0.01) for both H-154 and H-155 (r = 0.35, 0.37 and r = 0.48, 0.44, respectively) (Table 2). The MPH and BPH of HI were strongly and positively correlated with the MPH and BPH of TSW (r = 0.34 and 0.42, respectively) (P = 0.01) for H-154, but not for H-155. The MPH and BPH of HI were strongly and positively correlated with the MPH and BPH of BN and ISN (r = 0.35, 0.24 and r = 0.33, 0.25 at P = 0.01, respectively) for H-155, but not for H-154. The MPH of HI was negatively correlated with the MPH of SY (r = −0.27) (P = 0.05) for H-154, but not for H-155. No correlation was found between HI heterosis and the heteroses of the other traits (PH, IL, and SS) in either of the two hybrid groups.
Estimation of the genetic distance of parental lines
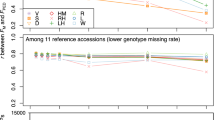
The GD of the parental lines was estimated using a total of 7600 SNPs (average major allele frequency >0.78 and gene diversity ranging from 0.19 to 0.29) in different B. napus linkage groups (Online Resource 4). Linkage group A10 had the fewest markers (182) with a marker density of one per 97.90 kb, while C03 had the most markers (915) with a marker density of one per 67.79 kb. Except for linkage groups A01, A04, A07, A10, C03, C05, and C07, more than 50 % of the SNPs in each linkage group had a polymorphism information content value over 0.20. The GD based on the SNPs between parental lines ranged from 0.20 to 0.60 for the H-154 group, and 0.15–0.50 for the H-155 group (Fig. 1; Online Resource 5). The GD between L-154 and L-155 was 0.47 (Online Resource 5). The GD values were unchanged when they were calculated using 35,791 SNPs selected for genome-wide association studies (Luo et al. 2015).
Correlations among GD, hybrid yield, and heterosis
The correlations among GD, hybrid yield, parental mean, and heterosis levels varied for seed yield and agronomic parameters (Table 3). For the H-154 and H-155 hybrids, GD was negatively and weakly correlated with hybrid yield for all traits, but was positively correlated with the MP of BY (r = 0.27) (P = 0.05) in the H-155 group. For heterosis, GD showed a positive and statistically significant correlation with the MPH and BPH of SY and BY (r = 0.25, 0.27 and r = 0.27, 0.29, respectively) (P = 0.05) in the H-154 group. No significant relationship was found between the difference in GD and the difference in heterosis levels of the traits in the two groups (Table 4). Additionally, significant negative correlations were observed between parental line yields and heterosis levels of hybrids for the most of the HI-related traits (P = 0.01 or P = 0.05), except the MPH and BPH of BY in the H-154 group, the BPH of BY in the H-155 group, and the BPH of BN in the H-155 group (Online Resource 6). The HI values for the parental lines were negatively correlated with the MPH and BPH of HI (P = 0.01) in the H-154 and H-155 groups (r = − 0.63, −0.51 and r = − 0.55, −0.48, respectively).
Discussion
Previous attempts to increase crop yields and avoid inbreeding depression had relied heavily on heterosis or hybrid breeding, for example, in maize (Duvick 2001), rye (Miedaner et al. 2002), sugar beet (Panella L, Lewellen R 2007), rice (Virmani 2012) and rapeseed (Gehringer et al. 2007; Ofori and Becker 2008). In the present study, all yield-related traits had high MPH and BPH, with widely variable values (Table 1). These results were consistent with those of previous studies (Qian et al. 2007; Xing et al. 2014), suggesting that HI possesses strong heterosis similar to other yield-related traits. Additionally, there is a possibility of improving HI in rapeseed through hybrid breeding. Furthermore, the mean values of most of the HI-related trait heteroses were significantly higher in H-154 than in H-155 (Online Resources 1 and 2), indicating that H-154 may be a promising parental inbred line for the production of elite hybrids in China.
Yield traits are complex, interrelated quantitative traits that are usually affected by genetics and the environment, as well as the interaction between these two factors. The total genetic (A and D) variance ratio was statistically significant for all traits (P = 0.01), ranging from 50 % (BY) to 87 % (PH and BN). The total genetic variance ratio for HI was 69 % and statistically significant. These results indicate that HI is mainly controlled by genetic effects (A and D), similar to other associated traits.
A total of 7600 SNPs distributed across 19 chromosomes were used to estimate the GD between the parental lines (Fig. 1; Online Resources 4 and 5) (Luo et al. 2015). As mentioned, the GD values were unchanged when calculated using 35,791 SNPs. Sang et al. (2015) used 40,201 loci to calculate the GD between parental lines and were unable to estimate the GD with fewer SNPs. However, our findings revealed that estimating the GD between parental lines can be achieved with considerably fewer markers.
In the present study, we first explored the relationship between GD and HI heterosis with high-throughput genomic SNPs, and found that GD was only correlated with the MPH and BPH of SY and BY in H-154 (Table 3). This suggested that GD is a valuable genetic parameter for predicting heterosis levels for these traits in the H-154 group. However, GD was not correlated with the HI of hybrids, the MP of parental lines, or HI heterosis (Table 3). The difference in the GD between parental lines was also not correlated with the difference in HI heterosis (Table 4). Which was inconsistent with the previous study, as seen in Sang et al. (2015). Inconsistent results have also been observed for traditional markers. Qian et al. (2007) found no significant correlations between heterosis and GD based on amplified fragment length polymorphisms. Xing et al. (2014) determined that GD based on simple sequence repeats was significantly correlated with the BPH of TSW. Additionally, parental line yield traits, including HI, were negatively correlated with their heteroses (Online Resource 6). These results reiterate that yield-related traits heterosis, including HI heterosis, are extremely complex. The efficiency of predicting heteroses with GD based on SNPs may be limited, even if the SNPs are an improvement over previous markers in terms of utility in high-throughput and accurate genotyping. Therefore, future studies will need to focus on the heterotic loci mapping with high-throughput genomic SNPs. These investigations may help to characterize the genetic mechanism of heterosis, which may aid in predicting heterosis in rapeseed.
References
Ali M, Copeland L, Elias S, Kelly J (1995) Relationship between genetic distance and heterosis for yield and morphological traits in winter canola (Brassica napus L.). Theor Appl Genet 91(1):118–121
Blair M, Cortés A, Penmetsa R, Farmer A, Carrasquilla-Garcia N, Cook D (2013) A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theor Appl Genet 126(2):535–548
Chen ZJ (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14(7):471–482
Diers B, McVetty P, Osborn T (1996) Relationship between heterosis and genetic distance based on restriction fragment length polymorphism markers in oilseed rape (Brassica napus L.). Crop Sci 36(1):79–83
Duvick DN (2001) Biotechnology in the 1930s: the development of hybrid maize. Nat Rev Genet 2(1):69–74
Ganal MW, Altmann T, Röder MS (2009) SNP identification in crop plants. Curr Opin Plant Biol 12(2):211–217
Gehringer A, Snowdon R, Spiller T, Basunanda P, Friedt W (2007) New oilseed rape (Brassica napus) hybrids with high levels of heterosis for seed yield under nutrient-poor conditions. Breed Sci 57(4):315–320
Hua J, Xing Y, Wu W, Xu C, Sun X, Yu S, Zhang Q (2003) Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci 100(5):2574–2579
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Liu R, Qian W, Meng J (2002) Association of RFLP markers and biomass heterosis in trigenomic hybrids of oilseed rape (Brassica napus × B. campestris). Theor Appl Genet 105(6–7):1050–1057
Luo X, Ma C, Yue Y, Hu K, Li Y, Duan Z, Wu M, Tu J, Shen J, Yi B (2015) Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genom 16(1):379
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9(5):356–369
Miedaner T, Gey AK, Sperling U, Geiger H (2002) Quantitative-genetic analysis of leaf-rust resistance in seedling and adult-plant stages of inbred lines and their testcrosses in winter rye. Plant Breed 121(6):475–479
Ofori A, Becker HC (2008) Breeding of Brassica rapa for biogas production: heterosis and combining ability of biomass yield. Bioenerg Res 1(1):98–104
Panella L, Lewellen R (2007) Broadening the genetic base of sugar beet: introgression from wild relatives. Euphytica 154(3):383–400
Qian W, Sass O, Meng J, Li M, Frauen M, Jung C (2007) Heterotic patterns in rapeseed (Brassica napus L.): i. Crosses between spring and Chinese semi-winter lines. Theor Appl Genet 115(1):27–34
Riaz A, Li G, Quresh Z, Swati M, Quiros C (2001) Genetic diversity of oilseed Brassica napus inbred lines based on sequence-related amplified polymorphism and its relation to hybrid performance. Plant Breed 120(5):411–415
Riddle NC, Birchler JA (2008) Comparative analysis of inbred and hybrid maize at the diploid and tetraploid levels. Theor Appl Genet 116(4):563–576
Rohlf F (2000) Multivariate analysis system, version 2.10 e. Applied Biostatistics. Inc, New York
Sang SF, Wang H, Mei DS, Liu J, Fu L, Wang J, Wang WX, Hu Q (2015) Correlation analysis between heterosis and genetic distance evaluated by Genome-Wide SNP Chip in Brassica napus. Sci Agric Sin 48(12):2469–2478
Shi J, Li R, Zou J, Long Y, Meng J (2011) A dynamic and complex network regulates the heterosis of yield-correlated traits in rapeseed (Brassica napus L.). PLoS One 6(7):371
Shull GH (1946) Hybrid seed corn. Science 103:547–550
Snowdon RJ, Iniguez Luy FL (2012) Potential to improve oilseed rape and canola breeding in the genomics era. Plant Breed 131(3):351–360
Teklewold A, Becker HC (2006) Comparison of phenotypic and molecular distances to predict heterosis and F1 performance in Ethiopian mustard (Brassica carinata A. Braun). Theor Appl Genet 112(4):752–759
Virmani SS (2012) Heterosis and hybrid rice breeding, vol 22. Springer Science & Business Media, Berlin
Xiao J, Li J, Yuan L, Tanksley SD (1995) Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140(2):745–754
Xing N, Fan C, Zhou Y (2014) Parental selection of hybrid breeding based on maternal and paternal inheritance of traits in rapeseed (Brassica napus L.). PLoS One 9:e103165
Yan J, Yang X, Shah T, Sánchez-Villeda H, Li J, Warburton M, Zhou Y, Crouch JH, Xu Y (2010) High-throughput SNP genotyping with the GoldenGate assay in maize. Mol Breed 25(3):441–451
Yu S, Li J, Xu C, Tan Y, Gao Y, Li X, Zhang Q, Maroof MS (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci 94(17):9226–9231
Yu C, Hu S, Zhao H, Guo A, Sun G (2005) Genetic distances revealed by morphological characters, isozymes, proteins and RAPD markers and their relationships with hybrid performance in oilseed rape (Brassica napus L.). Theor Appl Genet 110(3):511–518
Zhu J, Weir B (1994) Analysis of cytoplasmic and maternal effects I. A genetic model for diploid plant seeds and animals. Theor Appl Genet 89(2–3):153–159
Acknowledgments
This work was funded by the National Support Plan (2010BAD01B), the Central College Fund (2011PY155), and the National High Technology Research and Development Program of China (2011AA10A104, 2012AA101107).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, X., Ma, C., Yi, B. et al. Genetic distance revealed by genomic single nucleotide polymorphisms and their relationships with harvest index heterotic traits in rapeseed (Brassica napus L.). Euphytica 209, 41–47 (2016). https://doi.org/10.1007/s10681-015-1629-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1629-3