Abstract
Plants rely on RNA silencing (RS) as a major layer of innate defense to neutralize invading viruses. In an opposite way, plant viruses evolved a survival mechanism, based on viral suppressors of RS (RSS), able to hinder the host RS activation system. In the present study, the sequences of the barley yellow striate mosaic virus (BYSMV) proteins P3, P6, P9 and of Phosphoprotein (P), from Chahar Mahall (Iran) isolate, were determined. Furthermore, we carried out agroinfiltration assays in leaves of Nicotiana benthamiana (16c line) to explore the RS suppression mechanism of these proteins. Analyses showed that P and P3 act as weak local and limited short-term systemic inducers of RSS. A reduction of RNA-dependent RNA polymerase 6 (RDR6) gene expression was observed in presence of BYSMV P and P3 proteins, which imply their putative role in interfering with systemic RS, in which RDR6 is involved. In addition, P and P3 RSS activity were reduced when co-expressed with other BYSMV accessory proteins, indicating that, in the system used, only free P or P3 proteins lead to RSS. The BYSMV P protein lacked any detectable Glycine-Tryptophan (WG/GW) or F-box motifs, known to interfere with the AGO1 slicing activity. On the opposite, two WG/GW were identified in P3 sequence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Rhabdoviridae family includes a large group of viruses characterized by enveloped, bullet-shaped or bacilliform particles with a negative sense, single-stranded RNA genome. They infect a wide range of vertebrates, invertebrates, or plants, with a capacity to replicate in intermediate hosts. To date, plant rhabdoviruses are classified in four genera: Nucleorhabdovirus, Cytorhabdovirus, Dichorhavirus and Varicosavirus (Walker et al., 2018). The last two genera show a unique feature within the family, having a bipartite genome, with the polymerase included within a single RNA segment (Dietzgen et al., 2017).
Plant pathogenic rhabdoviruses infect both monocot and dicot hosts, including weeds and major crops such as rice, maize, wheat, potato, tomato, papaya, eggplant, lettuce, etc. They induce a variable symptomatology, ranging from latent infections with no visible symptoms, to stunting, yellowing, mosaic on leaves and chlorosis of systemically infected tissues, that can give rise to necrosis followed by the plant death (Almasi et al., 2010; Jackson et al., 2005).
Most plant rhabdoviruses rely for transmission on phytophagous insects, on which they exclusively depend for dissemination (Cao et al., 2018). Transmission by specific insect vectors occurs in a propagative manner, involving the virus multiplication inside the insect body, finally leading to transmission when the vectors feed on their hosts.
Barley yellow striate mosaic virus (BYSMV) is a member of the genus Cytorhabdovirus, the causal agent of the barley yellow striate mosaic disease (Almasi et al., 2010), is widely distributed worldwide, representing a potential risk for different cereal crops such as wheat, maize, rice, barley, oats, and triticale. It is transmitted by the plant hopper Laodelphax striatellus in a persistent, propagative manner (Cao et al., 2018). BYSMV infections provoke extensive crop losses particularly in wheat and millet fields in Middle East countries such as Iran. In 1989, an outbreak of BYSMV occurred in an experimental station causing severe reductions in yields (Izadpanah et al., 1991). Subsequently, Almasi et al., (2010) described and partially characterized an Iranian isolate of BYSMV. Data produced supported the closeness of this isolate to the northern cereal mosaic virus (NCMV). Yan et al. (2015) confirmed this proximity, by characterizing the complete genome of a BYSMV isolate from the Hebei province in northern China, where the virus was diagnosed in wheat fields.
BYSMV management mostly relies on early field detection, vector control and selection of resistant germplasm. Suitable agronomic practices may be adopted, such as eliminating weed reservoirs for the plant hopper vector, and avoiding the coincidence of cereals emergence with the vector spring migration. For an effective management program, it is important to characterize the factors underpinning BYSMV specificity and virulence. Moreover, knowledge of the functional activity that BYSMV genes induce in the defence systems of infected plants may be useful when selecting germplasm with enhanced tolerance or resistance levels.
The rhabdovirus monopartite genome structure presents a common organization with five canonical genes that encode from 3′ to 5′: a nucleoprotein (N), a phosphoprotein (P), a matrix protein (M), a glycoprotein (G) and a RNA-dependent RNA polymerase (L). This genomic region is flanked by 3′ leader and 5′ trailer sequences, stop and start transcription signals. The ORFs are separated by non-transcribed intergenic sequences, that give rise to capped and polyadenylated mRNAs (Bejerman et al., 2015; Dietzgen et al., 2017; Dietzgen et al., 2020). The structural N protein assembles the minus-sense genomic RNA with the P and the L proteins, yielding a ribonucleoprotein complex. The M protein has different functionalities, it is involved among other things in the condensation of the RNP complex, and transcription regulation. Protein P is highly phosphorylated, and constitutes a fundamental, non-catalytic cofactor of the polymerase L, performing important and different roles in viral replication (Dietzgen et al., 2020; Ivanov et al., 2011). In BYSMV the P protein interacts with the host plant deadenylase carbon catabolite repression 4 (CCR4) protein, that is recruited and utilized for binding into viroplasm-like bodies, enhancing viral replication (Zhang et al., 2020). The G and M proteins support the virus entry into the host cell, allowing the assembly and budding of new viral particles. The L protein carries out the enzymatic activities of transcription and virus replication, the RNA synthesis, 5′ end capping and 3′ end polyadenylation of the mRNA (Leyrat et al., 2010).
In addition to these five main genes, the rhabdoviruses genome encodes accessory proteins, either within new ORFs or included in the canonical ones (Walker et al., 2011). It is hypothesized that this feature confers plasticity to the rhabdovirus genomes, likely driving their evolution towards higher diversity and complexity, accompanied by the emergence of new traits. Most of the accessory proteins are localised between the P and M ORFs, and are identified with consecutive numbering (i.e. P3, P4, and so forth). A functional capability has been identified for P3, that presents similarities with the 30 K superfamily features of the viral movement protein (MP), as well as a RNA binding capacity. Trans-complementation studies showed that the P3 of rice yellow stunt rhabdovirus (RYSV) favoured the inter-cellular movement of a potato virus X (PVX) movement defective mutant (Huang et al., 2005). The P6 protein from RYSV has been recognised as a suppressor of RNA silencing (RS). Transient expression assays showed that P6 prevented the genesis of secondary siRNAs by blocking the RNA-dependent RNA polymerase 6 (RDR6), without impeding the local RS response, either in Nicotiana plants or rice protoplasts (Guo et al., 2013). In the case of alfalfa dwarf virus (ADV) and lettuce necrotic yellows virus (LNYV) the ability to perform a RS suppressor (RSS) activity resides in the P protein. Both viruses possess weak local RSS, differing during the systemic infection. Assays showed that ADV P protein holds a strong RSS activity, while studies carried out with LNYV P protein showed only a delay in the systemic spread of RSS (Bejerman et al., 2016; Mann et al., 2015). There is no evidence yet about the contribution of the additional genes in virus replication and survival. However, they seem to be involved in some unknown performance, as their current function is not yet completely defined.
The active competition for resources between plants and viruses gives rise, in most cases, to refined interaction mechanisms developed by both opponents. In this competition, plants express an RS defence pathway targeting the viral RNA to contrast infection (Guo et al., 2016). Invading viruses, in turn, are known to apply a counter-defence mechanism, evolved to avoid suppression by the host (Burgyán & Havelda, 2011). RS is a conserved mechanism present in all eukaryotes and acting in a sequence-specific manner (Csorba & Burgyán, 2016). It is involved in many cellular activities including plant physiology regulation and development or control of transposable elements (Xie et al., 2011). RS acts far beyond internal cell regulation, because it is very active also against external stimuli such as biotic or abiotic stresses, including plant defence from pathogens (Vance & Vaucheret, 2001).
RS is triggered by the formation of double stranded RNA (dsRNA) molecules that either arise endogenously or derive from the viral replication cycle. In brief, the viral dsRNA structure goes through different structural changes mediated by members of conserved host protein families. One of them is the ribonuclease type III-like enzyme named Dicer-like protein (DCL), in charge of slicing the dsRNA into 21–24 nucleotides small interfering RNA (siRNA) duplexes. Eventually, one strand of the duplex is guided by Argonaute (AGO) proteins to the degradation of the complementary viral strand. This mechanism relies on other proteins such as RDR6 that amplifies the signal throughout the plant and integrates all components in a highly coordinated regulatory pathway (Boccara et al., 2014; Chapman & Carrington, 2007). The dsRNAs produced by invading viruses elicit a silencing host defensive reaction, leading to the elimination of the pathogen RNAs. However, some viruses produce in turn some proteins that interfere with the host defensive reaction, modulating the pathogen RSS mechanisms (Cao et al., 2018). Several viral proteins targeting the RS plant response have been described, acting through different mechanisms. They include, among others: the potyviral helper component proteinase and viral genome-linked protein (HCPro, VPg), the cucumoviral 2b protein, the P38 protein of the turnip crinkle virus, the P25 protein of PVX, and the P19 protein of tombusviruses (Burgyán & Havelda, 2011). Similar to other plant viruses, rhabdoviruses own RSSs that counteract the host anti-viral mechanism, through the gene products of i.e. P proteins of LNYV and ADV (Bejerman et al., 2016; Mann et al., 2015) and P6 of RYSV (Guo et al., 2013). The P protein acts locally by inhibiting microRNA guided AGO1 cleavage, and systemically by repressing the production of secondary siRNAs (Bejerman et al., 2016; Mann et al., 2016). P6 acts systemically by interacting with RDR6 (Guo et al., 2013).
In the present study, we report about the molecular characterization of the phosphoprotein (P) and proteins P3, P6, and P9 of BYSMV Iranian isolate Chahar Mahal. Furthermore, we carried out agro-patch assays to gain a first insight about their possible role in the RSS mechanism.
Material and methods
Virus source and identification
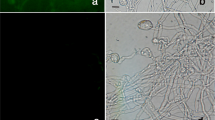
BYSMV was detected by PCR amplification reaction in a barley (Hordeum vulgare L.) sample exhibiting typical symptoms of barley yellow striate mosaic disease, including stunting and mosaic (Fig. 1a, b), collected in spring 2017 in the Chahar Mahall and Bakhtiari province (Iran). Total RNA was extracted from infected barley tissues using DENAzist Asia RNA extraction kit (DENAzist, Mashhad, Iran). The RNA was used as template in a reverse transcription - polymerase chain reaction (RT–PCR) assay using a specific primer pair targeting a portion of the viral glycoprotein gene, according to sequences available in GenBank (acc. KP163565 for BYSMV isolate Zanjan-1, Table 1). The reaction was carried out in a total volume of 20 μL, using 1 μg of RNA as template, 4 μL of 5× reaction buffer, 2 μL of dNTP mix (10 mM), 2 μL of reverse primer (400 nM), 1 μL of RevertAid™ Reverse Transcriptase (200 U), and 8 μL of Nuclease–free H2O. The mix was incubated at 42 °C for 1 h and then heated to 70 °C for 10 min to inactivate the enzyme. The PCR reaction was carried out in 25 μl containing a final concentration of 1 x PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs, 200 nM of both forward and reverse primers, 1 U Taq DNA polymerase and 2.5 μl of cDNA. The virus was also transmitted to and propagated in barley seedlings, as described by Almasi et al. (2010).
Symptoms of barley yellow striate mosaic virus (BYSMV) infection on barley plants showing leaves with striate yellow mosaic and necrosis along veins (a-b). Agarose gel showing amplified fragment (1200 bp amplicon) specific for BYSMV glycoprotein in leaves extracts from barley plants that developed systemic symptoms (lanes 1, 2, and 3). No amplification product was obtained from healthy plants, (lane 4). M = 1 kb ladder (Promega) (c). Amplified products of BYSMV ORFs of P3, P6, P9 and phosphoprotein P, with length of 516, 303, 156 and 888 bp, in lanes 1–4 respectively. M = 100 bp ladder (Promega) (d)
Plasmid constructs and bacterial strains
Transient expression assays were carried out to check the possible role of different BYSMV proteins in RSS, using a GFP reporter protein as an indicator of plant-induced RNA silencing. A high fidelity enzyme was used for PCR amplification of the P, P3, P6 and P9 ORFs from the Iranian BYSMV isolate. Specific primers were designed according to sequences from acc. KP163566, proceeding from isolate Zanjan (for P), and acc. KM213865 from strain Hebei, for P3, P6 and P9 ORFs, respectively (Table 1). The forward and reverse primers included recognition sites for NcoI and BamHI, respectively. In addition, a Hemagglutinin (HA) tag sequence was introduced at the N-terminal region for protein detection. The cDNA was used to amplify the specific targets using the Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Sc., MA, USA), according to the manufacturer recommendations, with the following thermal profile: 94 °C for 30 s, followed by 35 cycles at 94 °C for 30 s and 60 °C for 30 s, with a final step of polymerization at 72 °C for 5 min.
The plasmids used for the transient expression assays are based on pCKGFP (Reichel et al., 1996) and were modified by replacing GFP with the EGFP coding sequence (Carluccio et al., 2014, 2018). Amplified fragments were cloned first in pGemT vector properly polyadenylated and then sub-cloned in pCKEGFP, to finally be transferred to pCAMBIA for plant agro-infiltration. The pCKEGFP has the GFP gene driven by the constitutive 2x35S promoter from cauliflower mosaic virus and translator enhancer. This plasmid was digested with NcoI and BamHI that remove the GFP, and then properly dephosphorylated, to be subsequently substituted by the target gene of interest (GoI) under study. The entire cassette of the recombinant plasmid was then transferred to the pCAMBIA expression binary vector, digested by PSTI and appropriately dephosphorylated. The plasmids obtained were used for the transformation of Agrobacterium tumefaciens through the freeze-thaw method. The DH5-α Escherichia coli strain was used for DNA cloning and replication, and A. tumefaciens strain C58C1 was used for plant agro-infiltration assays. Cloned fragments corresponding of all constructs were used for DNA sequencing in both directions using an external service (Macrogen Europe, Amsterdam, NL).
Plant material and transient expression assay
Plants of Nicotiana benthamiana transformed line 16c, were grown under controlled conditions at 24 ± 2 °C with a photoperiod of 16 h light/8 h dark, until the 5–6 leaves stage. Single colonies of A. tumefaciens harbouring the transformed plasmid, with the appropriate BYSMV fragment constructs, were inoculated in 5 ml LB liquid medium supplemented with antibiotics. pCAMBIA with an empty plasmid (EV) and an HC-Pro isolated from PVYc-to (Mascia et al., 2010) were used as negative and positive controls, respectively. The broth cultures were incubated overnight at 28 °C with constant shaking and then used to inoculate 50 ml of fresh broth, and allowed to grow for 24 h more, under the same conditions. Bacterial sediments were obtained by centrifugation at 4 °C at 4000 rpm for 15 min, washed once with 10 mM MgCl2 and then re-suspended in infiltration buffer (10 mM MgCl2, 10 mM MES and 0.2 mM of acetosyringone). The bacterial suspension was normalised to OD600 = 0.15 and incubated for 3–4 h at room temperature. Co-infiltration assays were carried out by mixing equal volumes of A. tumefaciens cultures carrying the recombinant pCAMBIA with the BYSMV respective gene and the pCAMBIA-GFP plasmid. The leaves of N. benthamiana were agro-infiltrated into the abaxial side with 1 ml of bacterial suspension. The plants were checked at 2, 5, 14, and 21 days post-infiltration (dpi) for visualisation of local and systemic plant responses, monitoring the fluorescence levels with a hand-held long-wave UV lamp. Images were recorded using a Canon EOS700D digital camera and digitally contrasted.
RNA extraction, quantitative real-time PCR and northern blot analysis
Total RNA was extracted from 30 to 50 mg of agro-infiltrated leaf tissues with TRI Reagent (Sigma), and then treated with Turbo DNA-free kit (Invitrogen™), according to the manufacturer’s instructions. For first strand synthesis, total RNA from each sample was used with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™, CA, USA). The cDNA synthesis reaction was carried out in a total volume of 20 μL. From each template sample, 1 μg of total RNA was incubated at 65 °C for 5 min, placed on ice, then adding the following reagents: 2 μL of 10X RT-buffer, 0.8 μL of 25X dNTP mix (100 mM), 2 μL of 10X Random Primers, 1 μL MultiScribe™ reverse transcriptase, 1 μL RNase inhibitor and 3.2 μL Nuclease-free H2O. The mix was incubated for 10 min at 25 °C, followed by 120 min at 37 °C and the reaction inactivated by heating at 85 °C for 5 min.
The quantitative RT-PCR (qRT-PCR) assays were carried out by using as a normalizer the housekeeping gene encoding a serine-threonine phosphatase 2A protein (PP2A), an ubiquitously expressed member of the PPP gene family, with specific primer pairs for the green fluorescent protein (GFP), RDR6, and AGO1 genes, respectively (Table 2). qRT-PCRs were performed using a StepOne Plus™ Real-Time system (Applied Biosystems), under the following cycling conditions: 95 °C for 20 s, and 40 cycles at 95 °C for 3 s and 60 °C for 30 s, using 2× Fast SYBR™ Green PCR master mix. All qRT-PCRs were carried out in three biological and technical replicates. The fold expression of target mRNAs was analysed over the reference values. The results were calculated by eq. 2-DDCt, where DCt = specific Ct of target – Ct of internal control, and DDCt = control sample Ct – DCt of each experimental sample (Livak & Schmittgen, 2001).
Northern blot hybridization of normalized total RNA was carried out as described in Mascia et al. (2010), using a complementary Dig-labelled probe to sense GFP gene, synthesized through an in vitro transcription reaction in presence of the UTP digoxigenin labelled (DIG)-11-UTP. The membrane was hybridized overnight following the instructions for the RNA-RNA DIG hybridization protocol. The hybridized fragments were detected in a chemiluminescent assay, according the manufacturer’s recommendations.
Protein analyses
Proteins were isolated from the red, lower organic layer of TRI Reagent® (Sigma), after homogenizing the samples for RNA extraction, according to the manufacturer’s protocol. Protein extracts were analysed after running a 12% SDS- PAGE. The gel was blotted to a PVDF membrane with Trans-Blot® Turbo™ System (Bio-Rad, CA, USA). For GFP blot detection the primary rabbit anti-GFP antibody (Ab) was used, followed by the goat anti-mouse IgG (H + L) Horseradish Peroxidase (HRP) conjugated secondary Ab. For the HA tagged BYSMV proteins, the primary mouse anti-HA Ab and Goat anti-Mouse IgG, with (H + L) HRP conjugated secondary Ab were used, in presence of clarity western ECL substrate (Bio-Rad). Finally, film-based images were recorded. For detection of low molecular weight proteins, a 10% Tricine gel was used according to the protocol of Schägger and von Jagow (1987). Precision plus protein kaleidoscope 10–250 K was used as protein size marker. A gel run in the same conditions as for antibody detection was used to verify equal loading, and analysed by Coomassie brilliant blue staining.
Statistical and phylogenetic analyses
In the present study 10 to 25 plants, at the 5–6 leaves stage, were used per treatment. Four leaves per plant were co- infiltrated with the different constructs, in addition to the HC-Pro positive control of RSS and the pCAMBIA-GFP binary vector used as negative RSS. The data were subjected to analysis of variance (ANOVA) with subsequent Tukey’s means comparison using PAST (Paleontological Statistics Software Package) (Hammer et al., 2001). Pairwise analyses were carried out using the Poisson correction model. Evolutionary analyses were conducted with MEGA X (Kumar et al., 2018) using the Neighbour-joining method for the phylogenetic position of the analyzed fragments.
Results
Molecular characterization of BYSMV proteins
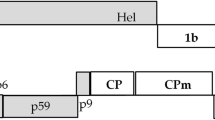
The presence of BYSMV in barley samples from southwest Iran was confirmed using a specific primer pair targeting the highly conserved glycoprotein gene (8/G), that yielded a fragment of the predicted size (1200 nt, Fig. 1c). The virus was successfully transferred to healthy plants via L. striatellus vector as described by Almasi et al. (2010). The infected plants were then used for RNA extraction, and the subsequent synthesis of cDNA from total RNA allowed the amplification of BYSMV ORFs encoding P, P3, P6 and P9 proteins (Fig. 1d). Their nucleotide sequences were determined and deposited in GenBank. The P gene (888 nt) encodes a putative phosphoprotein of 295 aa, (acc. MH509196). P3 (516 nt) encodes a protein with 171 aa (acc. MH509194). P6 and P9 are small ORFs flanking the M - G genes, of 303 and 156 nt long, respectively, (acc. MH509195, MH509197). The deduced aa sequence of the putative BYSMV P was aligned with other sequences within members of Rhabdoviridae (pairwise comparison, Table 2). Deduced aa sequence of ORF6 from the Iranian isolate was compared to those from other already described BYSMV isolates. P6 pairwise protein alignment showed an identity of 58% and 41% when compared with BYSMV (YP_009177227) and maize yellow striate virus (ATN96439) respectively, denoting a great sequence variability. ORF P9 and ORF P3 showed highest similarity to BYSMV Hebei isolate, with 100% identities at nucleotide and aa level for ORF P9, and 97.5 and 99% identities for ORF P3, respectively. NJ phylogenetic tree based on P nucleotide sequences confirmed affinity of the BYSMV Iranian isolate with the Hebei and Zanjan ones (Fig. 2).
Phylogenetic tree constructed by comparing the nucleotide sequences of the phosphoprotein P of members of Family Rhabdoviridae: barley yellow striate mosaic virus [BYSMV] - Iran isolate (* = this study), BYSMV- Zanjan isolate, BYSMV-Hebei isolate, BYSMV-SJZ-3 isolate, maize yellow striate virus [MYSV], maize sterile stunt virus [MSSV], northern cereal mosaic nucleorhabdovirus [NCMV], papaya cytorhabdovirus isolate Los Rios_Ec, rice stripe mosaic virus [RSMV], bean-associated cytorhabdovirus strain BaCV-BR-GO, Cytorhabdovirus sp. Isolate [SoyFace-2], cabbage cytorhabdovirus-1 strain FERA_050726, yerba mate virus A isolate Gob. Virasoro, persimmon virus A isolate persimmon, maize mosaic nucleorhabdovirus [MMV]. With the exception of NCMV and MMV (underlined) all the other members belong to Cytorhabdovirus genus. The tree was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units of the evolutionary distances used to infer the phylogenetic tree (evolutionary analyses carried out with MEGA X)
Agroinfiltration of BYSMV P and accessory ORFs to assess RSS activity
An agro-patch bioassay was carried out to evaluate the RSS activity of P3, P6, P9 and P proteins. The single genes were cloned in the binary vector pCAMBIA, under the Cauliflower mosaic virus 35S promoter. Transformed A. tumefaciens (strain C58C1), carrying the recombinant plasmids, were delivered into leaves of N. benthamiana line 16c by agroinfiltration. The transient expression assays considered the co-infiltration of two cultures, one containing the binary vector pCAMBIA with the GFP reporter, and the second one with the binary vector carrying the single BYSMV genes, respectively (Fig. 3). Furthermore, another assay was carried out agroinfiltrating the BYSMV gene combinations: P + P3 (P/P3), P + P6 (P/P6), P + P9 (P/P9) and finally all the proteins together (P/P3/P6/P9). Co-infiltration of Agrobacterium strains harboring the PVY helper component of isolate c-to (HC-Pro), or an empty vector, were used as positive and negative controls of RSS, respectively. The agro-patch assay was monitored for local fluorescent GFP expression at 2 and 5 dpi under UV light, and by a Western blot protein assay.
Schematic representation of BYSMV genome organization, according to Yan et al., 2015, showing core genes (light green arrows), and ancillary genes (dark green arrows) (a). BYSMV ORFs (single dark arrows) replaced the GFP, one at a time, into the expression cassette of the pCAMBIA vector (b). P = phosphoprotein; 3, 6, and 9 correspond to auxiliary proteins P3, P6 and P9, respectively
At 2 dpi, the infiltrated patches displayed a strong green fluorescence-signal in all trials, in comparison with the leaves of N. benthamiana 16c line, without any treatment (Fig. 4 panel a, 2 dpi). The plant response was in line with a high expression of the gene constitutively present in the 16c line, along with the agro-infiltrated pCAMBIA-GFP.
Leaves of Nicotiana benthamiana (line 16c) co-infiltrated with a 1:1 mixture of Agrobacterium tumefaciens carrying the plasmid pCAMBIA-GFP reporter and pCAMBIA with the different recombinant constructs, of BYSMV proteins (a, c). HC-Pro was used as positive control, being a strong suppressor of gene silencing, thus protecting the agro-infiltrated GFP. An empty vector (EV) was used as negative control. HC-Pro produced a strong GFP fluorescent signal, as it counteracted the GFP-silencing response of the plant. The negative control shows instead an active RS mechanism, where the GFP is degraded, yielding a red spot. Viral insertions: P (phosphoprotein) and P3, P6, and P9 alone, and also combinations P/P3, P/P6, and P/P9 to test for protein synergy. Fluorescence was recorded 2 and 5 dpi under UV light. GFP protein accumulation was determined by Western blots of infiltrated tissues at 2 and 5 dpi, using an anti-GFP antibody (PL = bands showing protein loading as control) (b, d). All samples were loaded with the same amount of leaf extract, except HC-Pro (first lane) due to the saturation of its band signal
At 5 dpi the GFP fluorescence was higher in the patches infiltrated with the HC-Pro, indicating an active RSS as expected. The BYSMV P protein agro-infiltrated leaves showed instead a decline in the GFP signal, although it was still possible to detect the GFP fluorescence, even if at a lower extent. A similar pattern was found in presence of the P3 ancillary gene, even alone or in combination with P protein, although at reduced level (Fig. 4a, 5 dpi). The agro infiltration with the GFP vector alone showed an evident red spot, a symptom of the active plant defense RS mechanism (Fig. 4a, 5 dpi, EV), as the presence of the GFP induced RS of the cognate gene, giving rise to the red patch. We observed that the P/GFP and P3/GFP assays were more effective in local RSS than the other tested constructs carrying P6 and P9, alone or in combination. Co-infiltrated leaves with GFP/P6 and P9 showed a pale fluorescence decline, higher than the negative control but lower when compared to the positive test or the other proteins, likely to be a consequence of a very mild RSS activity. The GFP protein accumulation was assessed by western blot analysis of infiltrated leaves, with the pCAMBIA-GFP plasmid together with BYSMV constructs singly or in combination, with the anti-GFP antibody (Fig. 4b and c, at 2 and 5 dpi), confirming the presence of the GFP protein in all assays. We checked the proteins in all assays and all were expressed.
According to these results, the interaction of the P protein with the individual BYSMV auxiliary proteins P6 and P9 had no positive effect on the local RSS, in which fluorescence and protein accumulation was lower, when compared with the P/P3 association.
Transient expressions of the single tested BYSMV constructs were confirmed at 2 and 5 dpi by Western blot analyses using the anti-HA flag-tagged antibody (Fig. 5a, 2 and 5 dpi). The detection assay highlighted bands of the expected molecular weights in all the treatments, corresponding to P3 (~ 18.6 kD), P6 (~ 10.8 kD), and P9 (~ 6.3 kD). In the case of the P (~ 33.3 kD), a further product of higher molecular weight was present at 5 dpi in addition to the band of the expected size (data not shown), likely due to a tendency of P to oligomerize (Liu et al., 2018) as shown by different blot assays conditions on 10% Tricine gel (data not shown).
BYSMV proteins in leaves of Nicotiana benthamiana 16c line co-infiltrated with Agrobacterium tumefaciens mixtures of pCAMBIA-GFP reporter and pCAMBIA recombinants carrying BYSMV P, (phosphoprotein) and P3, P6, and P9 (auxiliary proteins), alone. The anti-Ha-flag tagged antibody was used for detection of the transient expression of the BYSMV proteins at 2 and 5 dpi (a, top = protein detection, bottom = protein loading, PL). Northern blot analysis (b) of total RNA from samples shown in Fig. 4, at 5 dpi (GFP and HC-Pro or GFP and empty-vector, EV, as the positive and negative controls of RSS). BYSMV proteins are P, P3, P6, and P9, P/P3, P/P6, and P/P9. The blot was hybridized with a full length GFP probe. RNA equal loading was verified by gel red stained (rRNA)
Moreover, the level of this local mild RSS was tested by the accumulation at 5 dpi of the GFP-specific transcript as shown by a northern blot hybridization assay (Fig. 5b). Total RNA was extracted from infiltrated leaves at 5 dpi for detection of the GFP mRNA with a full-length GFP RNA probe generated by in vitro transcription from a DNA template, using the DIG-UTP system. The hybridization signal showed a higher accumulation of the GFP-specific transcript for the P and auxiliary P3 protein-infiltrated leaves, even if not at the same extent of the positive control. Treatments in which the P6 and P9 proteins were tested alone, and their different combinations, showed at 5 dpi very weak bands (Fig. 5b), with the GFP mRNA accumulation almost undetectable in their leaf patches. These data indicate that the P and the P3 protein of BYSMV have a local RSS activity visible at 5 dpi, not evident, however, in the P/P6, P/P9, and P/P3/P6/P9 associations.
The treated plants were monitored until 21 dpi. Complete systemic silencing was observed in upper non-infiltrated leaves of the 16c line with the pCAMBIA-GFP/empty-vector, where no green fluorescence was observed under UV light (Fig. 6, EV). In contrast, the pCAMBIA-GFP/HC-Pro treatment showed a bright green fluorescence evident in the whole plants (Fig. 6, HC-Pro). Except for GFP/P and GFP/P3 BYSMV, alone or in combination, all the other treatments showed progress of RS, as recorded by the rate of systemic silenced plants in the different treatments (Table 3).
Nicotiana benthamiana 16c leaves at 21 dpi, agro-infiltrated with HC-Pro, pCAMBIA-GFP/empty-vector (EV) and pCAMBIA-GFP/BYSMV constructs, P, P3, P6, P9, P/3, P/6, P/9 and all proteins P/P3/P6/P9, respectively. The plants (%) that showed systemic silencing suppression (out of the total tested) is shown in Table 3
Real time qPCR analysis
The silencing suppression potentiality of BYSMV ORFs was evaluated following the GFP expression in the agro-infiltrated leaves, RT-qPCR assays were carried out with RNA samples collected from the 5 dpi assays. In the GFP/P and the GFP/P3 assay the GFP transcript showed a higher expression when compared to the agro-infiltrated negative control and the others protein tested, alone or in combination. These data further support the hypothesis that those proteins confer a weak local RSS activity (Fig. 7a). In this assay we could not compare the GFP expression with the strong RSS HC-Pro of PVY because the use of a strong suppressor did not allow monitoring of the possible effects of weak suppressors. In fact, the RSS exerted by HC-Pro yielded a very high fluorescence level that could hide the other assays (data not shown).
qRT-PCR analyses showing GFP mRNA levels in discs of Nicotiana benthamiana 16c leaves agro-infiltrated with empty-vector (EV) and BYSMV construct, P, P3, P6, P9, P/P3, P/P6, and P/P9 at 5 dpi (a). The GFP transcript quantification is relative to the levels measured on control 16c (EV) plants and normalized by the expression of PP2A gene. Expression levels of AGO1 (b) and RDR6 (c) in agro-infiltrated leaves with P, P3 and P/P3 of BYSMV, and empty-vector (EV) at 5 dpi. Vertical bars show the standard error. Asterisks show statistically significant differences, at p < 0.05 (*) and p < 0.01 (**), respectively. Expression of GFP in plants co-infiltrated with HC-Pro was >18 folds those of the other samples (data not shown)
The analysis carried out on the expression of AGO1, and RDR6 during the co-infiltration assays of pCAMBIA-GFP/P or GFP/P3 and the combination of both proteins, showed a decrease of expression of both genes with significant differences according to analysis of variance (P < 0.05) and Tukey’s pairwise comparisons (Fig. 7b, c).
Discussion
Viruses of higher organisms evolved different defense mechanisms to neutralise host antagonism. This arms race likely gave rise to different strategies to override plant RS. More than one molecule able to act as plant suppressor, of different evolutionary origins, have been described in plant viruses (Burgyán & Havelda, 2011), which evolved to play an important role in the virus survival. ADV P, a homologue of the BYSMV P protein, is also a weak local RSS able, however, to restrict systemic RS (Bejerman et al., 2016). Data produced by Mann et al. (2015) on LNYV P revealed a capacity of the virus in limiting local RS with a mild RSS activity, deferring plant systemic RS. Other than Cytorhabdovirus, other viruses such as sonchus yellow net virus (SYNV), a Nucleorhabdovirus member, showed a capacity of the viral P protein to induce a local RSS (Jackson & Li, 2016). In the case of SYNV and ADV, only a weak local RSS activity was observed (Bejerman et al., 2016), similar to that of BYSMV isolated P protein herein described. BYSMV P and P3, alone or in combination, showed a weak local and systemic silencing activity, whereas P3 having a higher capability. P6 and P9 gave no sign indicative of a RNA silencing suppression. The GFP mRNA levels in plants co-infiltrated with pCAMBIA-GFP/P and pCAMBIA-GFP/P3, separately or in combination, showed the same trend, with a higher transcript accumulation. The combination of both proteins, however, did not display a synergistic RSS effect.
The ADV P3 protein has been described as a putative MP, a protein supporting viral genome spread through plasmodesmata, a fundamental process for systemic infection. Trans-complementation studies also confirmed its involvement in the cell-to-cell passage of Cyto- and Nucleorhabdovirus (Bejerman et al., 2015; Mann et al., 2016). Several studies delineated a further role of MP as viral enhancers of RSS, in which the suppressor of RNA silencing is indispensable for short- and long-term viral movements (Bayne et al., 2005). Amari et al. (2012) described a particular feature during the transient expression of MP, in which an enlargement of the silenced cells was observed, with an edge encircling the agro patch, considered as a mechanism exerted to enhance plant silencing signal spread. This phenotypic trait was also observed in this study, with the P and P3 agro infiltrated leaves presenting a red rim around the infiltrated zone that was more evident in P (Fig. 4a chart P, 5 dpi). The genomes of plant viruses have a restricted number of genes that encode essential proteins, their evolution favouring energy savings by holding different functionalities in a single gene. In this perspective, MP is in charge either of cell-to-cell and long-distance movements, including a defence mechanism counteracting the plant RS. An interaction between P and P3 in ADV has been reported by Bejerman et al. (2015), suggesting that multiple viral functions are dwelling in P3.
BYSMV P6 and P9 ORFs encode accessory proteins with unknown functions. While P9 showed 100% identity with BYSMV Hebei isolate at aa level, P6 disclosed only a 58% identity (aa) with the same isolate. This is indicative of a wide genetic variability or recombination within BYSMV field isolates. Differing from another rhabdoviruses, the BYSMV P6 protein + GFP did not block the GFP silencing, indicating that the protein was unable to suppress RS. The P6 of RYSV is effective in inhibiting the systemic spread of the plant silencing signal, although it is unable to interfere with the local RS. Guo and co-workers (2013) assayed a heterologous system, with PVX as vector for expression of RYSV P6, leading to an increase of virulence in N. benthamiana. Moreover, RYSV P6 also impaired the function of RDR6 in the tested plants. In this study, co-infiltration assays with the GFP, and P6 or P9 of BYSMV, showed no RS inhibition, either local or systemic. Nevertheless, BYSMV P6 showed a quite weak RSS activity, that was displayed as a slight time-delayed plant response. The combinations of BYSMV proteins lead to the obliteration of the local activity. Mann et al. (2015) showed that cytorhabdovirus LNYV free P protein worked better alone in local RSS than when combined with N or P3 (M) proteins, known to act in a synergistic mode. These results were also observed for BYSMV P3, which like the single P, had a better RSS inhibitory effect when expressed alone (Fig. 4a-d).
A weak systemic RSS activity of BYSMV P and P3 proteins was revealed in N. benthamiana 16c. Bejerman et al. (2016) showed that the ADV P protein has a weak local and a strong systemic RSS activity. ADV P6, however, did not display a local nor systemic suppression activity.
Different studies pointed out an important role of RDR6 in plant defense against several pathogens, in particular viruses and viroids (Boccara et al., 2014; Guo et al., 2013; Hong et al., 2015). It hence represents an important target of the viral RSS mechanism. The qRT-PCR of RDR6 displayed a relative gene expression decrease in leaf patches agroinfiltrated with P and P3 constructs. The transcript level was 3.0 and 2 times lower than the RSS control (16c + GFP) in P and P3, respectively. This suggests that both proteins interfere with the antiviral RS pathway. Hong et al. (2015) showed a decrease of RDR6 transcripts in rice after rice dwarf phytoreovirus (RDV) infection, with an increase of plants susceptibility.
AGO1 is an RNaseH-like enzyme incorporated in the RNA-inducing silencing complex (RISC). This enzyme is required for proper functioning of the RS machinery. It is partially responsible for the selection of the guiding strand, that is paired with the target and induces degradation of the complementary “passenger” strand (Schott et al., 2012). AGO1 orchestrates the RS plant defence, being highly transcribed under viral infection. The expression in P and P3, alone or in combination, was significantly lower than in 16c + GFP samples (Fig. 7b). RSS acts at the translational level, controlling the protein that is down regulated during viral infection (Carbonell & Carrington, 2015). In our study the mRNA of AGO1 in N. benthamiana resulted down regulated at 5 dpi in P and P3 assays, indicating a different AGO1 gene expression pattern. This observation fits data reported by Tahmasebi et al. (2017), concerning a reduction of AGO1 transcripts in plants agroinfiltrated with the suppressor of potato virus A. Várallyay and Havelda (2013) detected no AGO1 alteration during the N. benthamiana infection with tobacco etch virus (TEV), highlighting the occurrence of different plant responses to RSS likely depending on the “type” of virus suppressor own, a prevailing situation also observed in this study. Viral suppressor proteins have been reported to act through AGO inhibition at the translational level. They include the P38 protein of turnip crinkle virus (TCV), as well as the P1 of sweet potato mild mottle virus (SPMMV). In SPMMV the P1 protein has a WG/GW motif at the amino terminal region that is involved in AGO1 binding, resulting thus important in the RSS performance. In ADV P no GW/WG or F box motifs occur, suggesting that the protein uses alternative sites for AGO 1 binding (Bejerman et al., 2016). In other viruses, such as SPMMV, the P1 protein interacts directly with AGO1 (Almasi et al., 2015; Giner et al., 2010). A computational assay to identify possible WG/GW Argonaute-binding domains, performed at http://bioinfo.amu.edu.pl/agos/ using Agos (Zielezinski & Karlowski, 2011), showed no WG/GW motif in the BYSMV P, whereas two motifs were found in P3, suggesting that the former likely relies on a different RSS action mechanism.
Plant rhabdoviruses own singular silencing suppressors to counteract hosts RS, as this function has been identified in different molecules. Our data recognise for the first time in BYSMV a putative RSS function for P and P3 proteins. Assays herein described identify a possible function of these two additional BYSMV proteins, although more information is needed to check their performance. Data suggest that, within plant Rhabdoviridae, different genes with varying RSS capability are likely present. Furthermore, the function of BYSMV P6 did not match the RSS activity described for other rhabdoviruses, denoting a different but still unknown performance, in this species, for this protein.
References
Almasi, R., Afsharifar, A., Niazi, A., Pakdel, A., & Izadpanah, K. (2010). Analysis of the complete nucleotide sequence of the polymerase gene of barley yellow striate mosaic virus- iranian isolate. Journal of Phytopathology, 158, 351–356.
Almasi, R., Miller, W. A., & Ziegler-Graff, V. (2015). Mild and severe cereal yellow dwarf viruses differ in silencing suppressor efficiency of the P0 protein. Virus Research, 208, 199–206.
Amari, K., Vazquez, F., & Heinlein, M. (2012). Manipulation of plant host susceptibility: An emerging role for viral movement proteins? Frontiers in Plant Science, 3–10.
Bayne, E. H., Rakitina, D. V., Morozov, S. Y., & Baulcome, D. C. (2005). Cell-to-cell movement of potato Potexvirus X is dependent on suppression of RNA silencing. Plant Journal, 44(3), 471–482.
Bejerman, N., Giolitti, F., de Breuil, S., Trucco, V., Nome, C., Lenardon, S., & Dietzegen, R. G. (2015). Complete genome sequence and integrated protein localization and interaction map for alfalfa dwarf virus, which combines properties of both cytoplasmic and nuclear plant rhabdoviruses. Virology, 483, 275–283.
Bejerman, N., Mann, K. S., & Dietzgen, R. G. (2016). Alfalfa dwarf cytorhabdovirus P protein is a local and systemic RNA silencing suppressor which inhibits programmed RISC activity and prevents transitive amplification of RNA silencing. Virus Research, 224, 19–28.
Boccara, M., Sarazin, A., Thiebeauld, O., Jay, F., Voinnet, O., Navarro, L., & Colot, V. (2014). The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP-and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathogens, 10(1), e1003883.
Burgyán, J., & Havelda, Z. (2011). Viral suppressors of RNA silencing. Trends in Plant Science, 16(5), 265–272.
Cao, Q., Xu, W. Y., Gao, Q., Jiang, Z. H., Liu, S. Y., Fang, X. D., Gao, D. M., Wang, Y., & Wang, X. B. (2018). Transmission characteristics of barley yellow striate mosaic virus in its planthopper vector Laodelphax striatellus. Frontiers in Microbiology, 9, 1419.
Carbonell, A., & Carrington, J. C. (2015). Antiviral roles of plant ARGONAUTES. Current Opinion in Plant Biology, 27, 111–117.
Carluccio, A. V., Zicca, S., & Stavolone, L. (2014). Hitching a ride on vesicles: Cauliflower mosaic virus movement protein trafficking in the endomembrane system. Plant Physiology, 164(3), 1261–1270. https://doi.org/10.1104/pp.113.234534.
Carluccio, A. V., Prigigallo, M. I., Rosas-Diaz, T., Lozano-Duran, R., & Stavolone, L. (2018). S-acylation mediates Mungbean yellow mosaic virus AC4 localization to the plasma membrane and in turns gene silencing suppression. PLoS Pathogens, 14(8), e1007207. https://doi.org/10.1371/journal.ppat.1007207.
Chapman, E. J., & Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nature Reviews Genetics, 8(11), 884–896.
Csorba, T., & Burgyán, J. (2016). Antiviral silencing and suppression of gene silencing in plants p1–33 In: Wang, A., Zhou, X. (eds), Current research topics in plant virology. Springer International Publishing.
Dietzgen, R. G., Kondo, H., Goodin, M. M., Kurath, G., & Vasilakis, N. (2017). The family Rhabdoviridae: Mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Research, 227, 158–170.
Dietzgen, R. G., Bejerman, N. E., Goodin, M. M., Higgins, C. M., Huot, O. B., Kondo, H., Martin, K. M., & Whitfield, A. E. (2020). Diversity and epidemiology of plant rhabdoviruses. Virus Research, 281, 197942.
Giner, A., Lakatos, L., García-Chapa, M., López-Moya, J. J., & Burgyán, J. (2010). Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathogens, 6(7), e1000996.
Guo, H., Song, X., Xie, C., Huo, Y., Zhang, F., Chen, X., Geng, Y., & Fang, R. (2013). Rice yellow stunt rhabdovirus protein 6 suppresses systemic RNA silencing by blocking RDR6-mediated secondary siRNA synthesis. Molecular Plant-Microbe Interactions, 26(8), 927–936.
Guo, Q., Liu, Q., Smith, N. A., Liang, G., & Wang, M. B. (2016). RNA silencing in plants: Mechanisms, technologies and applications in horticultural crops. Current Genomics, 17, 476–489.
Hammer, Ø., Harper, D. A. T., & Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4(1), 1–9.
Hong, W., Qian, D., Sun, R., Jiang, L., Wang, Y., Wei, C., Zhang, Z., & Li, Y. (2015). OsRDR6 plays role in host defense against double-stranded RNA virus, Rice dwarf Phytoreovirus. Scientific Reports, 5, 11324.
Huang, Y. W., Geng, Y. F., Ying, X. B., Chen, X. Y., & Fang, R. X. (2005). Identification of a movement protein of rice yellow stunt rhabdovirus. Jornal of Virolology, 79(4), 2108–2114.
Ivanov, I., Yabukarski, F., Ruigrok, R. W., & Jamin, M. (2011). Structural insights into the rhabdovirus transcription/replication complex. Virus Research, 162(1–2), 126–137.
Izadpanah, K., Ebrahim-Nesbat, F., & Afsharifar, A. R. (1991). Barley yellow striate mosaic virus as the cause of a major disease of wheat and millet in Iran. Journal of Phytopathology, 131(4), 290–296.
Jackson, A. O., & Li, Z. (2016). Developments in plant negative-Strand RNA virus reverse genetics. Annual Review of Phytopathology, 54, 469–498.
Jackson, A. O., Dietzgen, R. G., Goodin, M. M., Bragg, J., & Deng, M. (2005). Biology of plant rhabdoviruses. Annual Review of Phytopathology, 43, 623–660.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549.
Leyrat, C., Gérard, F. C., de Almeida Ribeiro Jr., E., Ivanov, I., Ruigrok, R. W., & Jamin, M. (2010). Structural disorder in proteins of the rhabdoviridae replication complex. Protein and Peptide Letters, 17(8), 979–987.
Liu, Y., Du, Z., Wang, H., Zhang, S., Cao, M., & Wang, X. (2018). Identification and characterization of wheat yellow striate virus, a novel leafhopper-transmitted nucleorhabdovirus infecting wheat. Frontiers in Microbiology, 9, 468.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods, 25(4), 402–408.
Mann, K. S., Johnson, K. N., & Dietzgen, R. G. (2015). Cytorhabdovirus phosphoprotein shows RNA silencing suppressor activity in plants, but not in insect cells. Virology, 476, 413–418.
Mann, K. S., Bejerman, N., Johnson, K. N., & Dietzgen, R. G. (2016). Cytorhabdovirus P3 genes encode 30K-like cell-to-cell movement proteins. Virology, 489, 20–33.
Mascia, T., Cillo, F., Fanelli, V., Finetti-Sialer, M. M., De Stradis, A., Palukaitis, P., & Gallitelli, D. (2010). Characterization of the interactions between cucumber mosaic virus and potato virus Y in mixed infections in tomato. Molecular Plant-Microbe Interactions, 23(11), 1514–1524.
Reichel, C., Mathur, J., Eckes, P., Langenkemper, K., Koncz, C., Schell, J., Reiss, B., & Maas, C. (1996). Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proceedings of the National Academy of Sciences of the United States of America, 93(12), 5888–5893.
Schägger, H., & von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry, 166(2), 368–379.
Schott, G., Mari-Ordoñez, A., Himber, C., Alioua, A., Voinnet, O., & Dunoyer, P. (2012). Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. The European Molecular Biology Organization Journal, 31(11), 2553–2565.
Tahmasebi, A., Afsharifar, A., Rabiee, S., & Izadpanah, K. (2017). Altered expression of autophagy-related genes in Nicothiana benthamiana plants in response to potato virus a HC-pro silencing suppressor. Iranian Journal of Plant Pathology, 53(1), 63–74.
Vance, V., & Vaucheret, H. (2001). RNA silencing in plants--defense and counterdefense. Science, 292, 2277–2280.
Várallyay, E., & Havelda, Z. (2013). Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Molecular Plant Pathology, 14(6), 567–575.
Walker, P. J., Dietzgen, R. G., Joubert, D. A., & Blasdell, K. R. (2011). Rhabdovirus accessory genes. Virus Research, 162(1–2), 110–125.
Walker, P. J., Blasdell, K. R., Calisher, C. H., Dietzgen, R. G., Kondo, H., Kurath, G., Longdon, B., Stone, D. M., Tesh, R. B., Tordo, N., Vasilakis, N., Whitfield, A. E., & ICTV Report Consortium. (2018). ICTV report consortium ICTV virus taxonomy profile: Rhabdoviridae. Journal of General Virology, 99(4), 447–448.
Xie, W., Liang, C., & Birchler, J. A. (2011). Inhibition of RNA interference and modulation of transposable element expression by cell death in Drosophila. Genetics, 188(4), 823–834.
Yan, T., Zhu, J. R., Di, D., Gao, Q., Zhang, Y., Zhang, A., Yan, C., Miao, H., & Wang, X.-B. (2015). Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology, 478, 112–122.
Zhang, Z. J., Gao, Q., Fang, X. D., Ding, Z. H., Gao, D. M., Xu, W. Y., Cao, Q., Qiao, J. H., Yang, Y. Z., Han, C., Wang, Y., Yuan, X., Li, D., & Wang, X. B. (2020). CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication. Elife, 24, 9.
Zielezinski, A., & Karlowski, W. M. (2011). Agos-a universal web tool for GW Argonaute-binding domain prediction. Bioinformatics, 27(9), 1318–1319.
Acknowledgements
We thank D. Baulcombe for providing seeds of N. benthamiana transgenic line 16c and Dr. Livia Stavolone (IPSP, CNR) for kindly supplying the binary vector.
Availability of data and material
The material used for this publication is available upon request to the authors.
Funding
This work was financially supported by the Center of excellence in Plant Virology, Research and Technology, College of Agriculture, Shiraz University, Iran, as part of the first author PhD requirements.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The research does not involve any human participants and/or animals.
Consent for publication
All authors have reviewed the final version of this manuscript.
Conflict of interest
The authors declare that they do not have any conflict of interest.
Additional information
Accession numbers
GenBank accession numbers MH509194, MH509195, MH509196, and MH509197 were assigned to BYSMV_ P3 gene, BYSMV_ P6 gene, BYSMV_ Pp gene, and BYSMV_ P9 gene respectively.
Rights and permissions
About this article
Cite this article
Rabieifaradonbeh, S., Afsharifar, A. & Finetti-Sialer, M.M. Molecular and functional characterization of the barley yellow striate mosaic virus genes encoding phosphoprotein, P3, P6 and P9. Eur J Plant Pathol 161, 107–121 (2021). https://doi.org/10.1007/s10658-021-02307-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02307-7