Abstract
Interveinal chlorosis and leaf margin wrinkling are widespread symptoms of Cannabis sativa. They are traditionally attributed to the so-called hemp streak virus (HSV), but its existence has not been demonstrated yet. To our knowledge, no molecular investigation has so far been performed in order to identify the causal agent of this symptomatology, we therefore decided to use traditional and molecular virology techniques to better characterize symptoms and pursue the etiological agent. No pathogenic virus was found by using targeted PCR reactions and by RNA sequencing, whereas we were able to detect the Cannabis cryptic virus (CanCV) with both techniques. We, therefore, developed an RT-qPCR assay based on a CanCV-specific TaqMan probe and applied it to a wide range of symptomatic and symptomless plants, using a two-step (for quantification), or a one-step (for fast detection) protocol. Both symptoms and the virus were only shown to be transmitted vertically and did not pass via mechanical inoculation or grafting, though we could not find any cause-effect correlation between them. In fact, the virus was found in all the tested hemp samples, and its abundance varied greatly between different accessions and individuals, independently from the presence and severity of symptoms. The suggestion that hemp streak is caused by a virus is therefore questioned. Some abiotic stresses seem to play a role in triggering the symptoms but this aspect needs further investigation. For breeding purposes, a selection of parental plants based on the absence of symptoms proved to be efficient in containment of the disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cannabis sativa L. (Cannabaceae), commonly known as hemp, is an important herbaceous species and a uniquely versatile plant that has been grown for at least 5000 years (McPartland et al. 2000). This plant offers raw material for many industrial uses, including textiles, paper, rope and innovative biomaterials, as well as foodstuffs, cosmetics and fuel. It is also a source of bioactive molecules of interest to the pharmaceutical industry, but its use for human health has experienced ups and downs throughout history, and nowadays it is regulated differently in many countries.
Hemp is well adapted to most European agricultural and environmental conditions but, despite a reputation for being quite resistant, it can host several pathogens and suffer from diseases and pests (McPartland 1996). A number of known bacterial and fungal pathogens are responsible for different stress symptoms and diseases (Kusari et al. 2013), whilst limited literature on the subject reports only a dozen common plant viruses able to infect Cannabis plants (McPartland et al. 2000) and only five viral syndromes. Among these, three are caused by known agents, all with a worldwide distribution and a wide host range: Alfalfa mosaic virus (AMV), Cucumber mosaic virus (CMV) and Arabis mosaic virus (ArMV), whereas the causal agents of the syndromes observed and described under the names of Hemp streak virus (HSV) and Hemp mosaic virus (HMV) have yet to be isolated and characterized. In particular, symptoms attributed to HSV were originally described in 1941 (Röder 1941) and caused serious losses also in Italy (Ferri 1963). According to these references, foliar symptoms appear as interveinal yellow streaks; eventually, leaf margins become wrinkled, leaf tips roll upward and leaflets curl into spirals, and such syndrome is still widely observed among hemp accessions, as reported by farmers and amateur growers.
More recently, another virus was found in hemp plants and characterized by molecular tools. The Cannabis cryptic virus (CanCV) was accidentally isolated by Ziegler et al. (2012) while using hemp as host to propagate a hop virus. The CanCV genome consists of two linear molecules of monocistronic double-stranded RNA (dsRNA) 2420 and 2346 nucleotides in length, which code for a putative RNA-dependent RNA polymerase (RdRp) and a coat protein (CP), respectively. Following phylogenetic analysis, the virus has been included in the new Betapartitivirus genus, within the Partitiviridae family, whose members are known to infect plants, fungi, and protozoa generally without causing obvious symptoms (Nibert et al. 2014). Ziegler et al. (2012) found that five out of six hemp varieties tested were positive for the presence of CanCV by using CP specific primers in RT-PCR, suggesting that this virus is widespread among hemp varieties. Nonetheless, no other information is reported in literature about the role and effects of CanCV in Cannabis plants.
The recent resurgence of interest in both medical and food uses of C. sativa is demanding plants that are suitable for vegetative propagation and free from pathogens, like viruses, that can affect plant metabolism and production. Up to date, little is known about hemp diseases and very little research has been done, probably also due to regulatory constraints in its cultivation. However, a better understanding of mechanisms triggering recurring symptoms, like those of hemp streak, is essential for disease management and breeding of resistant varieties.
During this project, we studied expression and transmissibility of hemp streak symptoms and looked for the possible viral agent by means of both traditional and molecular techniques; these were applied according to each result achieved as work progressed. Corroborating evidence obtained by different techniques (for example RT-PCR and NGS) is included to strengthen our results, describing methods that can be readily used by other researchers. We also focused on CanCV, investigating its spread and transmissibility, therefore offering a new contribution to the controversial question on the attribution of an etiological role of ‘cryptoviruses’ in plants.
Materials and methods
Plant material
Different varieties of Cannabis sativa were used, as specified in the following paragraphs. Most experiments were carried out on the Dutch cultivar (cv.) ‘Chamaeleon’ and on the Italian variety ‘Codimono’ because, based on our experience, these varieties showed severe HSV associated symptoms (as defined in the following paragraph) starting from very early stages of growing. Also many other European and Asian accessions were assessed to investigate the diffusion of CanCV.
Plants were grown in three different environments. Most experiments were performed in a glasshouse where plants received natural photoperiod and lighting; a second environment consisted of a growth chamber equipped with high-pressure sodium lamps, with a photoperiod of 16 h of light and a temperature of 22–26 °C. The third environment was the open field in the experimental station of Rovigo, Italy (Latitude: 45° 4′ 41.912″ N; longitude: 11° 45′ 56.988″ E; altitude: 0 m asl).
Nicotiana benthamiana and Chenopodium quinoa plants were used as common viral hosts for transmission tests by mechanical inoculation.
Hemp streak symptom description
Early foliar symptoms attributed to HSV, as described by McPartland et al. (2000), were considered. These are typically interveinal yellow streaks (later called interveinal chlorosis) and leaf margin wrinkling upward (later referred to as leaf wrinkling). Severity scales for interveinal chlorosis and leaf wrinkling were defined from 1 (no symptoms) to 4 (symptoms spread on the whole leaf surface) (Fig. 1). In all experiments, plant phenotyping was performed independently by three different operators and average scores were calculated. The severity index (SI) of symptoms was given as the product of the values obtained for interveinal chlorosis and leaf wrinkling, ranging from 1 (no symptoms) to 16.
Screening for plant viruses
Leaves from 6 highly symptomatic (SI = 16) and 6 symptomless (SI = 1) plants of the cv. ‘Chamaeleon’ were tested for the presence of the most common viruses by PCR. The species or genus-specific primer pairs and relative references are listed in Table 1. DNA or cDNA were used as templates for amplification, according to the virus type. DNA was extracted from 50 to 100 mg of frozen leaf material with Nucleospin® Plant II (Macherey-Nagel) and 5 ng were used for subsequent applications.
Total RNA was extracted from 50 to 100 mg frozen leaf material by Spectrum™ Plant Total RNA kit (Sigma-Aldrich) and treated with DNAse I (Sigma-Aldrich). For cDNA synthesis, 500 ng of RNA were denatured at 80 °C for 2 min, then reverse transcribed in 10 μL reaction, using High-Capacity RNA-to-cDNA Kit (Life Technologies). For PCR detection of members of the genus Carlavirus and Potyvirus, the cDNA was synthesized using the M4T primer (Table 1) and the enzyme Superscript III (Invitrogen).
Positive controls (Table 1) were nucleic acids extracted from virus-infected plants. Fragments of the expected sizes were obtained by (RT)-PCR with all primer pairs used on the respective positive controls, supporting the reliability of the results obtained.
The presence of virus particles in symptomatic hemp leaves was further tested by electronic microscopy. Purification was performed as published elsewhere (Turina et al. 2007) from 60 g of symptomatic leaves collected from 2-month-old hemp plants. Purified particles were adsorbed to two different carbon-coated (polyvinyl formal) 300 mesh-grids and observed by Philips CM 10 transmission electron microscope (TEM) after negative staining with 2% uranyl acetate (pH 4.3).
Next generation sequencing approach
Total RNA from highly symptomatic cv. ‘Codimono’ was used as template for an RNA sequencing approach. The nucleic acid was extracted using QIAGEN RNeasy kit, followed by selective depletion of ribosomal RNA (Invitrogen RiboMinus™ Plant Kit). A paired-end library was created from double stranded cDNA (Illumina Nextera XT Library Preparation Kit) and sequenced on an Illumina MiSeq platform. De novo assembly of the reads was done using the Geneious software (ver. 9.2.3). The assembled contigs were analyzed for similarities to known viruses by NCBI nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify putative (partial) genomes of viruses present in the sample.
CanCV re-sequencing and sequence analysis
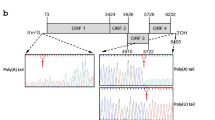
The entire genome of the CanCV isolate from the hemp cv. ‘Chamaeleon’ was re-sequenced as follows: the dsRNA was isolated from symptomatic leaves as described by Tzanetakis and Martin (2008), run on 1% agarose gel, and used as template for a random RT-PCR approach (Froussard 1992) and RACE reactions for the determination of 5′ and 3′-ends; remaining gaps were amplified using sequence specific primers. With the exception of RACE PCR products, which were sequenced directly, obtained fragments were cloned (pGEM-T vector, Promega) before sequencing (Helmholtz Centre for Infection Research, Braunschweig). Sequence analysis and alignments were carried out using BLAST, SMART (http://smart.embl-heidelberg.de) and EMBL-EBI Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) tools. The sequences obtained from the two genomic dsRNAs were submitted to GenBank (accession no. KX709964 and KX709965). The CP complete coding sequence (cds) was aligned to that of the CanCV isolate from the French variety Fedora 17 (GenBank ID: JN196537). The portion of CP gene between primers CP_F and CP_R (Table 2) was amplified by PCR from the cDNA of 5 accessions from different countries and sequenced in order to verify the level of conservation. Sequencing was performed by BioFab Research s.r.l (http://www.biofabresearch.it/). Sequence alignments were performed with EMBL-EBI Clustal Omega. CanCV CP and RdRp cds were blasted against the ‘Purple Kush’ and ‘Finola’ genome and transcriptome (van Bakel et al. 2011) and all the Cannabis ESTs available on NCBI (http://www.ncbi.nlm.nih.gov), in order to verify whether CanCV sequences were already present in these databases. Attempts of amplifications from genomic DNA of ‘Chamaeleon’ and ‘Codimono’ were also performed to exclude integration events into the plant genome.
All primer and probe sequences used in this work are listed in Table 2.
CanCV relative quantification
CanCV relative quantification was performed through two-step RT-qPCR using the Rotor-Gene 6000 Instrument (Corbett Life Science). TaqMan primers and probe (Table 2) were designed on a conserved region of CanCV CP by using PRIMER EXPRESS Software version 2.0 (Life Technologies) and blasted against the non-redundant nucleotide and EST collections (NCBI), and the Cannabis genome (http://genome.ccbr.utoronto.ca) to further confirm their specificity.
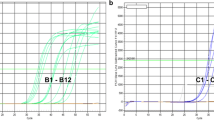
Each reaction was performed in a total volume of 15 μL, containing 7.5 μL of Rotor-Gene probe PCR kit 2X (Qiagen), 0.75 μL of TaqMan probe 20X (1X contains 900 nM of each unlabeled primer and 250 nM of the 6-FAM dye-labeled MGB probe) and 1.5 μL of cDNA as the template, obtained as described above. Amplifications were carried out with an initial activation step of 3 min at 95 °C, then 40 two-step cycles of 3 s at 95 °C and 10 s at 60 °C for annealing/extension.
Relative quantification was performed using beta tubulin TUB (Marks et al. 2009) as the reference gene in an independent 6-FAM labelled TaqMan reaction performed in the same run (Table 2). The absence of DNA in cDNA samples, obtained through DNase treatment of the starting RNAs before reverse transcription to cDNA, was further confirmed by PCR with intron-spanning primers targeting the TUB gene (Table 2) and agarose gel electrophoresis. Two standard curves were also generated to determine the amplification efficiency of both assays (CP and TUB). As templates, we used the PCR products of 525 bp (for CP) and of 293 bp (for TUB) obtained using primers as in Table 2, and purified on column by QIAquick PCR Purification Kit (Qiagen). The Ct values of 4 serial 1:100 dilutions of the templates were plotted versus the log of the dilution factors and the slopes of the linear regression curves were used to calculate efficiencies according to the eq. E = 10^(−1/slope)-1. Both efficiencies of amplification were close to 1 (1.01 for CP assay and 1.09 for TUB assay), data are thus presented as mean of ddCt ± standard error, calculated according to the rules of error propagation. For the CP assay, the standard curve loses linearity at Ct > 32, we therefore considered this Ct value as a threshold for a reliable relative quantification. Samples with Ct < 32 were CanCV positive (CanCV+) while CanCV was considered not quantifiable (CanCV_nq) if Ct ≥ 32 or when no amplification could be detected. These latter samples were double checked with an additional primer pair, targeting the RdRp cds of CanCV, designed by software Primer 3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi), and used with SYBR Green chemistry. Reactions were performed in 15 μL, containing 1X SYBR Select Master Mix (Life Technologies), 100 nM of specific primer pairs and 1.5 μL of cDNA template. Amplification conditions consisted of an initial denaturation at 95 °C for 2 min, followed by 45 cycles as follows: 95 °C for 15 s, 60 °C for 60 s. The absence of dimer formation was checked in no-template samples by product dissociation analysis and electrophoretic separation in agarose gel. Each analysis was repeated twice. In the event of a difference between the two replicates of 0.5 Ct or more the reactions were repeated.
For fast detection of CanCV, a spot one-step RT-qPCR method was also developed and used in the experiments on symptom and CanCV transmissibility. Templates prepared as in Minguzzi et al. (2016) were used in TaqMan reactions as described above, with the addition of 4 U of M-MLV Reverse Transcriptase (Promega) and 0.25 μL of RNasin Ribonuclease Inhibitor (Promega). The program was 30 min at 48 °C, 10 min at 95 °C, 40 cycles of 10 s at 95 °C and 1 min at 60 °C.
Inter and intra-accession analysis of CanCV
Seeds of 125 different hemp accessions were sown in the field in April 2013 and grown until maturity. Foliar symptoms were evaluated at pre-flowering (June 2013) and 35 single plants, derived from 12 different countries across Europe and Asia, with symptom severity ranging from SI = 1 to 16 were chosen to investigate the presence and the abundance of CanCV. A sample of the youngest fully expanded leaf (about 1 g, fresh weight) was collected at pre-flowering (June 2013), full flowering (July 2013) and end of flowering (August 2013), frozen immediately in liquid nitrogen and stored at −80 °C until RNA extraction. Two independent homogenization, RNA extractions and reverse transcriptions were performed on each leaf sample. Every cDNA was used as a template for two reactions of two step RT-qPCR as described above. Pairwise t-test and correlation coefficient were calculated with R.
In order to verify the reliability of the assay, data were analyzed with both the ddCt method, using TUB as the reference gene, and by absolute quantification, with the CP standard curve described above. In this case, the log of the viral copy number was estimated as in Olmos et al. (2005), which was modified to take into account that, in our case, the template is dsDNA, and normalized to the total RNA used in the RT-qPCR reaction. The two sets of results were rescaled and compared with the Two Sample Kolmogorov-Smirnov Test using R.
For intra-accession analyses, five different accessions were selected from the 35 field-collected accessions, covering a wide range of symptom severity and CanCV abundance. Five seeds per accession were sown in peat fertilized with 1X Hoagland’s solution 1 (Hoagland and Arnon 1950) and grown in a growth chamber with 16 h photoperiod, 430 μmol PAR, 23/19 °C day/night temperature and 65 ± 5% day/night relative humidity for 3 weeks. For each plant, the youngest fully expanded leaves were sampled and processed as described above.
Transmission tests
Symptoms were tentatively transmitted to plants of N. benthamiana and C. quinoa grown in a greenhouse for 3 weeks before mechanical inoculation. Leaf tissues from ‘Codimono’ plants showing severe symptoms were homogenized in a mortar with the addition of 0.1 M Na-phosphate buffer, pH 7.5 containing 0.12% sodium sulfite and 5% polyvinylpyrrolidone (Clover et al. 2003). The homogenate was mixed with carborundum powder and mechanically inoculated on herbaceous indicators (at least 10 plants for each species). The appearance of local or systemic symptoms was evaluated until 4 weeks after inoculation. This experiment was performed twice.
The same protocol was followed with the leaves of plants that were positive to CanCV. Local and systemic presence of CanCV in inoculated N. benthamiana and C. quinoa plants was evaluated by one-step RT-qPCR assay.
Cross grafting between rootstocks and scions from twelve symptomatic and twelve symptomless C. sativa (cv. Codimono) plants and from twelve plants that were positive to CanCV (CanCV+) and twelve others in which CanCV was not quantifiable (CanCV_nq) was also performed. All plants were grown in a growth chamber under the conditions described above and the symptoms, as well as the presence of CanCV, were monitored until 3 months after grafting.
The transmission of symptoms and CanCV was also evaluated in ‘Codimono’ progenies deriving from crosses of one symptomatic (SI =16) female CanCV_nq plant partially subjected to sex reversion with silver thiosulfate solution (STS) (Mohan Ram and Sett 1982) that was self-pollinated and used for pollinating three other female plants with the following characteristics: 1) symptomatic and CanCV+; 2) symptomless and CanCV+ and; 3) symptomless and CanCV_nq. Seeds were collected separately from each plant. Seed production, seed germinability, symptom rate in 40 plants, and CanCV presence in 3 symptomatic and 3 symptomless plants from the offspring were evaluated.
Finally, plants of the same ‘Codimono’ variety used in the previous test were rated for symptoms and then, at an early stage (17 days after sowing), divided into two groups consisting of 24 symptomatic (SI > =9) and 24 symptomless plants. These groups of plants were kept separate in grow boxes (1.2 m × 1.2 × 2.0 m) until seed production and the rate of symptoms was evaluated in 160 plants of the two progenies.
Effect of nutrition and growth conditions on symptoms
In order to verify the effect of nutrients upon the appearance of symptoms, seeds of ‘Chamaeleon’ were sown in February 2014 on coco coir or peat substrate. Plants were supplemented daily with the complete fertilizer Coco A & B (5% NO3−, 0.1% NH4+, 4% P2O5, 3% K2O, 7% CaO, 3% MgO, 2% SO3, 0.007% B, 0.001% Cu, 0.02% Fe DTPA, 0.0003% Fe EDTA, 0.01% Mn, 0.002% Mo, 0.007% Zn, 0.5% fulvic and humic acid, CANNA International BV, Oosterhout, Netherlands). Four different electrical conductivity (EC) levels of nutrient solution (0.3; 1; 2 and 3 mS/cm) were compared, approximately corresponding to a dilution of the complete fertilizer at 0, 1:500, 1:250, and 1:170 (v/v), respectively. Half of these were grown in a glasshouse with a natural photoperiod of approximately 14 h, and temperatures and humidity ranging as in Online Resource 1. The remaining plants were grown in the growth chamber with a 16-h photoperiod, 26/22 °C day/night temperature and 72/55% day/night relative humidity, and moved to the glasshouse 3 weeks after sowing. Five weeks after sowing, symptoms were rated and plant heights were measured.
Analysis of variance and the Tukey’s test post-hoc comparisons were performed with R.
Results
Screening for plant viruses
Different approaches were used to identify possible viral agents. Firstly, an approach based on PCR and specific primer pairs was attempted, comparing symptomatic and symptomless plants. The result was that no amplification was obtained with primers that are specific to viruses known for infecting Cannabis (CMV, AMV, ArMV). Negative results were also obtained using specific primers for both viruses with RNA (genera Trichovirus, Capillovirus and Foveavirus within the family Flexiviridae, Nepovirus group A, B and C, Tobamovirus, Potyvirus, Carlavirus and Luteovirus) and DNA (genera Caulimovirus and Badnavirus) genomes. Positive results were only obtained for CanCV that was detectable in both symptomatic and symptomless plants.
Subsequently, as a universal method to identify plant viruses, we performed an RNA-Seq analysis on a highly symptomatic sample, and again only CanCV was identified. In detail, chloroplast and mitochondrion DNA sequences were subtracted (accession nos. KR059940, KR779995, NC_026562, NC_027223 and NC_029855) from the 4,491,006 reads obtained from the Illumina MiSeq run. De novo assembly of the remaining 2,131,126 reads resulted in a total of 1000 assembled contigs, ranging from 232 up to 5949 bases. Only two contigs of 2380 and 2258 bases assembled from 18,975 and 11,475 reads, respectively, were identified as the putative genome (lacking the extreme 5′- and 3′-ends) of a Partitivirus, with the highest nucleotide sequence identity of 99% versus Cannabis cryptic virus RNA1 and RNA2 (isolate Fedora 17). No other contigs with similarities to known viruses or viroids could be identified.
Finally, TEM observation of the virus purification product identified isometric virus-like particles of approximately 34–36 nm in diameter, corresponding to the CanCV particles observed by Ziegler et al. (2012). No other type of virus-like structure was observed in any of the analyzed specimens.
CanCV database search and sequence analysis
A total of 20 CanCV CP sequences similar to CP cds from ‘Fedora 17’ were retrieved in the NCBI EST collection (EW700746, EW700777, EW700979, EW700980, EW701078, EW701242, EW701378, EW701434, EW701404, EW701449, EW701446, EW701462, EW701504, EW701513, EW701558, EW701651, EW701661, EW701691, EW701706), all derived from different cDNA libraries obtained from healthy, symptomless plants of the fiber hemp ‘Chamaeleon’ (van den Broeck et al. 2008).
No sequence similar to CanCV was retrieved from the Cannabis genome or amplified starting from hemp genomic DNA, suggesting the absence of integration events.
As expected, a single band of about 2.4 kbp resulted from agarose gel electrophoresis of the viral dsRNA extracted from the cv. ‘Chamaeleon’, which provided further evidence of the absence of other viruses with ssRNA or dsRNA genomes. Sequence analysis of the two genomic RNAs clearly identified the virus as an isolate of CanCV. The complete nucleotide sequences encoding the putative CP and RdRp genes were 99% identical to both the ‘Fedora 17’ isolate described by Ziegler et al. (2012), and the sequences obtained by deep sequencing of the hemp variety ‘Codimono’. This suggests a low sequence variability in all isolates for which sequence information is available.
The partial re-sequencing of CanCV CP from 5 different accessions further confirmed the presence of a highly-conserved portion which was therefore selected for the design of primers and probes for the TaqMan assay. The 65 bp region of the CanCV CP cds amplified with the TaqMan assay primers showed no similarity with other sequences. This supports the CanCV specificity of the assay.
CanCV relative quantification
Among the 125 phenotyped hemp accessions, approximately 74% were completely symptomless and 26% showed hemp streak symptoms on at least one plant. The CanCV relative quantification was performed on 35 single plants from different accessions, chosen on the basis of the SI. Nine were symptomless, 10 showed mild symptoms (2 < SI < 4) and the remaining plants showed more severe symptoms, with a SI ranging from 6 to 16. The results in Fig. 2 clearly showed and confirmed that CanCV is ubiquitous, with wide ranging quantities (the highest ddCt is about 24). The standard error is more or less constant across observations regardless of the mean, therefore for low ddCt the measurement is notably less precise. In our dataset, a difference in ddCt of 5 usually corresponds to a statistically significant difference according to pairwise t-test (p-value < 0.05), but the discrimination could be improved by increasing the number of replicates. The 35 accessions are displayed from left to right according to an increasing level of symptom severity (expressed as SI), making it clear that there is no direct correlation between CanCV relative quantity and symptoms (correlation coefficient = 0.14). Nor did the time of sampling influence the viral RNA quantity in hemp leaves (data not shown). Using the TaqMan assay, CanCV was found not quantifiable in two symptomless plants only, for which, however, the RdRp based SYBR Green assay revealed positivity to CanCV.
CanCV relative quantification in 35 plants of 35 different accessions. Plants, identified with a number from 1 to 35, are displayed from left to right according to increasing level of symptom severity, highlighted with different shades of gray. Accession 15 was used as the calibrator for ddCt calculation. Results are expressed as means of ddCt and error bars represent the standard errors. nq: accessions with non-quantifiable CP (Ct > 32)
These data were analyzed also by absolute quantification using the standard curve method and the distributions of the two sets of results were compared. The Two Sample Kolmogorov-Smirnov Test p-value of 0.9995 indicated that there is no significant difference between the two distributions, supporting the choice of using TUB as reference gene to estimate the relative quantification of the virus.
We also observed that the variability of CanCV quantification among plants of the same accession was generally very high (Table 3); a low variability of measures, associated to very high Ct values, was found only in one accession out of five (No. 15). Three plants of three different accessions were excluded from the analysis because their Ct was higher than the TaqMan assay quantification limit; however, for all three SYBR Green analysis unequivocally confirmed positivity to CanCV.
Transmission of symptoms and CanCV
No symptoms were observed on C. quinoa or N. benthamiana plants 4 weeks after mechanical inoculation, nor was CanCV detected in any plant inoculated with CanCV positive sap. Between 1 and 2 weeks after grafting, all scions were successfully grafted onto rootstocks, allowing translocation of nutrients and, if present, pathogens invading the vascular system. Despite symptomatic and CanCV-positive scions and rootstocks preserving their symptoms and viral infection, they transmitted neither of these to the symptomless or CanCV_nq scions and rootstocks up to 3 months after grafting.
Phenotyping and molecular analysis of progenies derived from vertical transmission experiments are shown in Table 4. Pollen was obtained from a symptomatic and CanCV_nq plant subjected to partial chemical-induced sex reversion. The highest rate of symptoms (52.5%) was registered in the self-pollinated progeny, whereas in the progenies derived from the other crosses symptoms did not seem to be clearly influenced by the presence or absence of symptoms on the female plants. Interestingly, CanCV was not quantifiable in the sexually reverted (symptomatic) plant, and progenies were clearly positive to CanCV when the female parental was positive, while Ct was higher than 32 (nq) when the female parental was nq too. Seed production of the symptomatic female was slightly lower than that of the symptomless ones (12 g vs 16 g and 19 g), while seed germinability was not clearly influenced by CanCV or by symptoms in the parents.
In the experiment where ‘Codimono’ plants were separated according to the presence or absence of symptoms at day 17, the selection of symptomatic plants provided highly symptomatic offspring (36.3%), while the selection of symptomless parents provided offspring with very low rate of symptoms (6.3%).
Effect of nutrition and growth condition on symptoms
The appearance and severity of symptoms were not triggered by nutritional deficiencies. Plants without fertirrigation (EC of 0.3 mS/cm) were strongly affected in their development, with an average height below 7 cm and presenting severe symptoms of nutrient deficiencies. At EC from 1 to 3 mS/cm, increasing levels of fertilization positively affected plant growth, but did not prevent the appearance of symptoms (Fig. 3) or decrease their severity (data not shown). On the other hand, the stress imposed by moving a batch of plants from a controlled environment to the glasshouse with no temperature, humidity or light control, where the difference between the lowest and highest temperatures often exceeded 20 °C and the photoperiod was reduced, triggered symptoms on about 20% of the plants (Fig. 3).
Fertilization and environmental effects upon symptoms. a percentage of symptomatic plants; b average height and standard errors of the mean (19 degrees of freedom). In gray: plants that were moved from the growth chamber to the glasshouse 3 weeks after germination; in black: plants that were in the glasshouse for the whole experiment. The electrical conductivity (EC) of the tested nutrient solutions was 0.3, 1, 2 and 3 mS/cm; peat was used as control. *: symptoms were not evaluated due to unsuitable plant growth. Different letters correspond to statistical difference according to Tukey’s post-hoc test (p < 0.05). No symptoms were observed at EC 2, 3 and on peat under glasshouse conditions
Discussion
Despite the traditional description of hemp streak as a viral disease, in our set of experiments it was not possible to identify any viral causal agent correlated to the symptoms.
Different approaches, based on both conventional and novel virology techniques, were consequently used, based on, and as confirmation of, the results obtained with previous methods. The initial approach was to test the presence of known viruses on symptomatic hemp plants, using PCR with specific primer pairs for as many viruses as possible, including, but not limited to those transmitted by pollen or seeds. By PCR analysis, we excluded the presence of viruses known to be pathogenic for C. sativa (AMV, CMV and ArMV) and of some other common plant viruses with RNA genomes (genera Trichovirus, Capillovirus, Foveavirus, Nepovirus, Potyvirus, Tobamovirus, Carlavirus, Luteovirus). Our analyses also failed to retrieve DNA viruses of the Caulimovirus and Badnavirus genera, which, to the contrary, can give consistent symptoms in other hosts. As a further attempt to identify never-before-seen viruses or other pathogens putatively involved in triggering symptoms, we therefore moved to an RNA next-generation sequencing approach, which offers a powerful alternative solution to conventional targeted methods (Adams et al. 2013). RNA virus genomes or pathogen transcripts are sequenced on a background of host nucleic acid, and identified by similarity to known pathogens using bioinformatic approaches. These innovative technologies are also a powerful tool in understanding the implications of one or more microorganisms in plant disease occurrence. In this research, we sequenced the transcriptome of highly symptomatic hemp leaves with Illumina MiSeq and the only match was with the Cannabis cryptic virus, which was also revealed by TEM. Indeed, CanCV was the only detectable virus in the accessions of Cannabis sativa we analyzed and therefore we investigated its spread and possible role in the syndrome under study. We started with a partial re-sequencing of the CP gene of CanCV isolated from different accessions to verify the conservation of its genome and, based on this, designed specific real-time PCR assays. We report two variants of the method of detection: the first enables measurement of the CanCV relative abundance in relation to that of the plant beta tubulin gene using RT-qPCR and TaqMan probes; the second is aimed at a non-quantitative fast diagnostic screening and it is based on the same CP primers and probes but in a single one-step Real Time RT-PCR using leaf juice as a template. CanCV was detected in all tested plants and our results demonstrated for the first time that CanCV abundance is extremely variable in both different accessions and different individuals of the same accession, but in a completely independent manner from hemp streak symptom appearance and severity.
Ziegler et al. (2012) had already reported a wide distribution of CanCV in hemp varieties, with only one accession (Uso 31) being tested negative. However, they used semi-quantitative PCR for detection, which is less sensitive than RT-qPCR with hydrolysis probes. We could not find any unequivocally negative samples, even though for some individuals the results were close to the limit of our detection techniques. Since partitiviridae sequences encoding for CPs have been found integrated into plant genomes (Chiba et al. 2011), we tested whether this event also occurred for CanCV in hemp, as a possible explanation of its ubiquitous presence. CP-like sequences were not found by searching in public genomic databases or by direct molecular analyses on plant DNA. Conversely, 20 ESTs similar to CanCV CP were retrieved from the cDNA libraries constructed by van den Broeck et al. (2008) from healthy plants, suggesting that the virus was present in the host plant at the time of sampling.
Since we did not find a cause-effect relationship between this virus and streak symptoms, further experiments would be necessary to unveil if and how CanCV affects the fitness of hemp plants. In our experimental conditions, the presence alone of CanCV did not trigger any symptoms and, at least in the tested genotypes, we can exclude the co-existence of more viral agents, that in other biological systems can cause a severe symptomatology, as reported in strawberry (Tzanetakis et al. 2008) and in many other species (Lamichhane and Venturi 2015). Ubiquitous presence and latent behavior are expected for these types of viruses that have been called cryptic precisely because they usually mediate persistent infections of their hosts, usually plants or fungi, with few or no effects on host cells. In some cases, they can negatively affect growth and virulence of some fungal hosts (Potgieter et al. 2013; Xiao et al. 2014), whereas, in other cases the same virus might be harmful, cryptic or beneficial to its host depending on the complexity of the ecological interaction (Hyder et al. 2013). The effect on plant pathogenic fungi can indirectly affect their interaction with the host plant. Some direct negative effects have been reported on host plants, i.e. on raphanus and beet (Chen et al. 2006; Xie et al. 1994), while other observations suggest positive mutualistic relationships, as in the interaction between White clover cryptic virus 1 and Trifolium repens or between Lolium perenne and a probable deltapartitivirus (Nibert et al. 2014; Roossinck 2011). In fact, plant viruses might positively impact response to both abiotic and biotic stresses (Roossinck 2015), but these relationships bring costs and benefits to the plant and the ability to understand and modulate these could be a powerful resource for breeders and growers. For all these reasons, monitoring the presence and abundance of CanCV in hemp could be of great interest. The availability of the TaqMan assays may be useful to underpin the biology and epidemiology of CanCV, which remain largely unknown, and in breeding programs in the attempt to identify virus-free selections. Our results also gave new evidence that CanCV is transmitted vertically at least by seeds, while it does not pass between scion and rootstock in the case of grafting.
Concerning the hemp streak symptoms, our results, according to the new Koch postulates (Fredricks and Relman 1996), suggested that leaf chlorosis and wrinkling are most likely not caused by a virus. Hemp streak symptoms were observed in many genotypes both in open field and in controlled conditions (growth chamber, glasshouse) since the appearance of the first mature leaves. We observed that in general leaf chlorosis and wrinkling appeared more intensely in young plantlets grown in stressful conditions (for example in rockwool pads). Abiotic factors like imbalances of soil nutrients and climatic stress can provoke symptoms that resemble those of diseases caused by living organisms or can predispose plants to infectious diseases caused by other agents (McPartland et al. 2000). Deficiency of Mg, Zn, Fe and Mn can provoke yellowing between veins but the association of this to the typical upward curling of leaf edges is difficult to attribute to a specific imbalance (lack or excess) of nutrients. Deficiency symptoms from mobile nutrients (like N, P, K, Mg, Mo) generally begin in larger leaves at the bottom of plants, while symptoms from less mobile elements (Mn, Zn, Ca, S, B, Fe and Cu) usually begin in younger leaves; however, we observed the appearance of symptoms at the same time on younger and older leaves. Moreover, the results obtained by our experiments excluded the possibility that nutritional deficiency alone could enhance symptom appearance and severity. Finally, we observed that sudden application of abiotic stresses, such as big changes in light, humidity and temperature, caused an outbreak of symptoms, while plants that germinated and grew in the same stressful environment seemed to adapt and barely showed symptoms; therefore the exact trigger still remains unknown. Controlled application of abiotic stresses could clarify their involvement, and this aspect would certainly merit further studies. Our results also confirmed that symptoms of leaf chlorosis and wrinkling are vertically transmitted as reported by McPartland et al. (2000), while they did not pass to herbaceous test plants or to symptomless C. sativa plants by grafting.
To conclude, the Hemp Streak syndrome has been observed for many years now, but the identification of its cause is still problematic. The presence of CanCV is not in itself synonymous with the disease, but we cannot exclude its interaction with other biotic or most likely abiotic stresses in triggering the symptomatology. Also the possible effect of other vertically transmitted characteristics, perhaps also of a genetic nature, would need further investigation. A better understanding of what initiates this syndrome would be helpful to deploy more appropriate control and management measures. However, since it is transmitted vertically, a selection of parental plants before crossing and, if possible, the control of environmental conditions, are effective in decreasing the rate of symptoms spreading to offspring.
References
Adams, I. P., Miano, D. W., Kinyua, Z. M., Wangai, A., Kimani, E., Phiri, N., et al. (2013). Use of next-generation sequencing for the identification and characterization of maize chlorotic mottle virus and sugarcane mosaic virus causing maize lethal necrosis in kenya. Plant Pathology, 62(4), 741–749. doi:10.1111/j.1365-3059.2012.02690.x.
Badge, J., Brunt, A., Carson, R., Dagless, E., Karamagioli, M., Phillips, S., et al. (1996). A carlavirus-specific PCR primer and partial nucleotide sequence provides further evidence for the recognition of cowpea mild mottle virus as a whitefly-transmitted carlavirus. European Journal of Plant Pathology, 102(3), 305–310. doi:10.1007/BF01877970.
Chen, J., Chen, J., & Adams, M. J. (2001). A universal PCR primer to detect members of the Potyviridae and its use to examine the taxonomic status of several members of the family. Archives of Virology, 146(4), 757–766. doi:10.1007/s007050170144.
Chen, L., Chen, J. S., Liu, L., Yu, X., Yu, S., Fu, T. Z., & Liu, W. H. (2006). Complete nucleotide sequences and genome characterization of double-stranded RNA 1 and RNA 2 in the Raphanus sativus-root cv. Yipinghong. Archives of Virology, 151(5), 849–859. doi:10.1007/s00705-005-0685-8.
Chiba, S., Kondo, H., Tani, A., Saisho, D., Sakamoto, W., Kanematsu, S., & Suzuki, N. (2011). Widespread endogenization of genome sequences of non-retroviral rna viruses into plant genomes. PLoS Pathogens, 7(7), e1002146. doi:10.1371/journal.ppat.1002146.
Chomič, A., Pearson, M. N., Clover, G. R. G., Farreyrol, K., Saul, D., Hampton, J. G., & Armstrong, K. F. (2010). A generic RT-PCR assay for the detection of Luteoviridae. Plant Pathology, 59(3), 429–442. doi:10.1111/j.1365-3059.2010.02282.x.
Clover, G. R. G., Pearson, M. N., Elliott, D. R., Tang, Z., Smales, T. E., & Alexander, B. J. R. (2003). Characterization of a strain of Apple stem grooving virus in Actinidia chinensis from China. Plant Pathology, 52(3), 371–378. doi:10.1046/j.1365-3059.2003.00857.x.
Digiaro, M., Elbeaino, T., & Martelli, G. P. (2007). Development of degenerate and species-specific primers for the differential and simultaneous RT-PCR detection of grapevine-infecting nepoviruses of subgroups A, B and C. Journal of Virological Methods, 141(1), 34–40. doi:10.1016/j.jviromet.2006.11.033.
Ferri, F. (1963). Alterazioni della canapa trasmesse per seme. Progresso Agricolo, 9, 346–351.
Foissac, X., Svanella-Dumas, L., Gentit, P., Dulucq, M.-J., Marais, A., & Candresse, T. (2005). Polyvalent degenerate oligonucleotides reverse transcription-polymerase chain reaction: a polyvalent detection and characterization tool for trichoviruses, capilloviruses, and foveaviruses. Phytopathology, 95(6), 617–625. doi:10.1094/PHYTO-95-0617.
Fredricks, D. N., & Relman, D. A. (1996). Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clinical Microbiology Reviews, 9(1), 18–33.
Froussard, P. (1992). A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Research, 20(11), 2900.
Hoagland, D. R., & Arnon, D. I. (1950). The water culture method for growing plants without soil. Barkley: The college of Agriculture, University of California Circular, 347.
Hyder, R., Pennanen, T., Hamberg, L., Vainio, E. J., Piri, T., & Hantula, J. (2013). Two viruses of Heterobasidion confer beneficial, cryptic or detrimental effects to their hosts in different situations. Fungal Ecology, 6, 387–396. doi:10.1016/j.funeco.2013.05.005.
Koziel, S. P. (2010). Genetic analysis of lignification and secondary wall development in bast fibers of industrial hemp (Cannabis sativa). Master’s Thesis in Plant Biology. University of Alberta. Retrieved from https://era.library.ualberta.ca/files/tt44pn501#.VkDHsNIvdpg.
Kusari, P., Kusari, S., Spiteller, M., & Kayser, O. (2013). Endophytic fungi harbored in Cannabis sativa L.: diversity and potential as biocontrol agents against host plant-specific phytopathogens. Fungal Diversity, 60(1), 137–151. doi:10.1007/s13225-012-0216-3.
Lamichhane, J. R., & Venturi, V. (2015). Synergisms between microbial pathogens in plant disease complexes: a growing trend. Frontiers in Plant Science, 6(May), 1–12. doi:10.3389/fpls.2015.00385.
Letschert, B., Adam, G., Lesemann, D. E., Willingmann, P., & Heinze, C. (2002). Detection and differentiation of serologically cross-reacting tobamoviruses of economical importance by RT-PCR and RT-PCR-RFLP. Journal of Virological Methods, 106(1), 1–10. doi:10.1016/S0166-0934(02)00135-0.
Lyttle, D. J., Orlovich, D. a., & Guy, P. L. (2011). Detection and analysis of endogenous badnaviruses in the New Zealand flora. AoB PLANTS, 11(1), 1–13. doi:10.1093/aobpla/plr008.
Marks, M. D., Tian, L., Wenger, J. P., Omburo, S. N., Soto-Fuentes, W., He, J., et al. (2009). Identification of candidate genes affecting Delta9-tetrahydrocannabinol biosynthesis in Cannabis sativa. Journal of Experimental Botany, 60(13), 3715–3726. doi:10.1093/jxb/erp210.
McPartland, J. M. (1996). A review of Cannabis diseases. Journal of the International Hemp Association, 3(9), 19–23 http://www.internationalhempassociation.org/jiha/iha03111.html.
McPartland, J. M., Clarke, R. C., & Watson, D. P. (2000). Hemp diseases and pests: management and biological control. New York: CABI Publishing.
Minguzzi, S., Terlizzi, F., Lanzoni, C., Poggi Pollini, C., & Ratti, C. (2016). A rapid protocol of crude RNA/DNA extraction for RT-qPCR detection and quantification of ‘Candidatus phytoplasma prunorum’. PLoS ONE, 11(1), e0146515. doi:10.1371/journal.pone.0146515.
Mohan Ram, H. Y., & Sett, R. (1982). Induction of fertile male flowers in genetically female Cannabis sativa plants by silver nitrate and silver thiosulphate anionic complex. Theoretical and Applied Genetics, 62(4), 369–375. doi:10.1007/BF00275107.
Nibert, M. L., Ghabrial, S. A., Maiss, E., Lesker, T., Vainio, E. J., Jiang, D., & Suzuki, N. (2014). Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Research, 188, 128–141. doi:10.1016/j.virusres.2014.04.007.
Olmos, A., Bertolini, E., Gil, M., & Cambra, M. (2005). Real-time assay for quantitative detection of non-persistently transmitted Plum pox virus RNA targets in single aphids. Journal of Virological Methods, 128, 151–155.
Pappu, H. R., & Druffel, K. L. (2009). Use of conserved genomic regions and degenerate primers in a PCR-based assay for the detection of members of the genus Caulimovirus. Journal of Virological Methods, 157(1), 102–104. doi:10.1016/j.jviromet.2008.11.014.
Potgieter, C. A., Castillo, A., Castro, M., Cottet, L., & Morales, A. (2013). A wild-type Botrytis cinerea strain co-infected by double-stranded RNA mycoviruses presents hypovirulence-associated traits. Virology Journal, 10(1), 220. doi:10.1186/1743-422X-10-220.
Röder, K. (1941). Einige Untersuchungen über ein an Hanf (Cannabis sativa L.) auftretendes Virus. Faserforschung, 15, 77–81.
Roossinck, M. J. (2011). The good viruses: viral mutualistic symbioses. Nature reviews. Microbiology, 9(2), 99–108. doi:10.1038/nrmicro2491.
Roossinck, M. J. (2015). Move over bacteria! Viruses make their mark as mutualistic microbial symbionts. Journal of Virology, 89, 6532–6535. doi:10.1128/JVI.02974-14.
Turina, M., Ricker, M. D., Lenzi, R., Masenga, V., & Ciuffo, M. (2007). A severe disease of tomato in the Culiacan area (Sinaloa, Mexico) is caused by a new picorna-like viral species. Plant Disease, 91(8), 932–941. doi:10.1094/PDIS-91-8-0932.
Tzanetakis, I. E., & Martin, R. R. (2008). A new method for extraction of double-stranded RNA from plants. Journal of Virological Methods, 149(1), 167–170. doi:10.1016/j.jviromet.2008.01.014.
Tzanetakis, I. E., Price, R., & Martin, R. R. (2008). Nucleotide sequence of the tripartite Fragaria chiloensis cryptic virus and presence of the virus in the Americas. Virus Genes, 36(1), 267–272. doi:10.1007/s11262-007-0186-9.
van Bakel, H., Stout, J. M., Cote, A. G., Tallon, C. M., Sharpe, A. G., Hughes, T. R., & Page, J. E. (2011). The draft genome and transcriptome of Cannabis sativa. Genome Biology, 12(10), R102. doi:10.1186/gb-2011-12-10-r102.
van den Broeck, H. C., Maliepaard, C., Ebskamp, M. J. M., Toonen, M. A. J., & Koops, A. J. (2008). Differential expression of genes involved in C1 metabolism and lignin biosynthesis in wooden core and bast tissues of fibre hemp (Cannabis sativa L.) Plant Science, 174(2), 205–220. doi:10.1016/j.plantsci.2007.11.008.
Wei, T., & Clover, G. R. G. (2008). Use of primers with 5′ non-complementary sequences in RT-PCR for the detection of nepovirus subgroups A and B. Journal of Virological Methods, 153(1), 16–21. doi:10.1016/j.jviromet.2008.06.020.
Xiao, X., Cheng, J., Tang, J., Fu, Y., Jiang, D., Baker, T. S., et al. (2014). A novel partitivirus that confers hypovirulence on plant pathogenic fungi. Journal of Virology, 88(17), 10120–10133. doi:10.1128/JVI.01036-14.
Xie, W. S., Antoniw, J. F., White, R. F., & Jolliffe, T. H. (1994). Effects of beet cryptic virus infection on sugar beet in field trials. Annals of Applied Biology, 124(3), 451–459. doi:10.1111/j.1744-7348.1994.tb04150.x.
Ziegler, A., Matoušek, J., Steger, G., & Schubert, J. (2012). Complete sequence of a cryptic virus from hemp (Cannabis sativa). Archives of Virology, 157(2), 383–385. doi:10.1007/s00705-011-1168-8.
Acknowledgements
The authors would like to thank Dr. Lanzoni Chiara, DipSA – Plant Pathology, University of Bologna, for technical support in the laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
The research leading to these results received funding from the European Union’s Seventh Framework Programme FP7-KBBE-2012-6-singlestage (collaborative project) for research, technological development and demonstration under grant agreement n° 311849. It was also supported by Italian projects RGV FAO of the Italian Ministry of Agriculture, Alimentation and Forest Policies (DM 29561 18/12/2014) and “Made in Italy - Alimenti funzionali: farine vegetali per cibi salutistici, ingradienti attivi ad elevata biodisponibilità” (DM 00028MI01 12/07/2011) of the Italian Ministry of Economic Development.
Electronic supplementary material
Online Resource 1
(DOC 65 kb)
Rights and permissions
About this article
Cite this article
Righetti, L., Paris, R., Ratti, C. et al. Not the one, but the only one: about Cannabis cryptic virus in plants showing ‘hemp streak’ disease symptoms. Eur J Plant Pathol 150, 575–588 (2018). https://doi.org/10.1007/s10658-017-1301-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-017-1301-y