Abstract
Over the last decade, comparative molecular profiling studies between compatible and incompatible plant-pathogen interactions have shown that susceptible response of the host to a pathogen requires factors that promote disease development. In this study, we examined proteome profiles during a compatible interaction between wheat and stripe rust. A 2D-LC system (ProteomeLab PF2D) was used for protein separation and to compare the proteome from infected and control samples. More than 700 protein peaks at each time point were compared between pathogen- and mock-inoculated samples. Selected proteins, with significant differences in abundance were identified by nanoLC-ESI- MS/MS and generated spectra were searched against the wheat protein databases from UniProt, and NCBI and the Puccinia database from The Broad Institute. In total, the identified proteins comprised of 62 % wheat and 38 % Pst proteins. All identified proteins were searched by bioinformatics-based algorithms to detect their subcellular localization and signal peptide motifs which have the potential to catch the candidate effector proteins. The wheat proteins were classified based on their function. Although a compatible interaction, many wheat proteins, such as antioxidants, PRs and cold-responsive proteins, are implicated in defense and stress tolerance. On the pathogen side, 64 proteins were identified, and included some important pathogenicity proteins that can play role in pathogen virulence and suppress the host defense. In addition, we discovered that nine proteins have a signal sequence and three of the hypothetical fungal proteins, PGTG_11681T0, PGTG_07231T0 and CBH50687.1, have been tentatively identified as candidate effectors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stripe rust (yellow rust), which is caused by the biotrophic fungal pathogen Puccinia striiformis f. sp. tritici (Pst), is one of the most damaging diseases affecting bread wheat worldwide, including Turkey. In wheat, major race-specific resistance genes (Yr) usually provide complete protection throughout the entire growth cycle of the plant. Therefore, the development of resistant wheat varieties that include more than one Yr gene is accepted as the most efficient, cost-effective and environmentally friendly strategy for controlling the disease. However, pathogens have a great potential to mutate and produce new strains that display enhanced virulence, spreading rapidly. These aggressive strains have caused the breakdown of resistance that is conferred by newly developed cultivars, which in turn cause severe pandemics. Therefore, more effective and rapid disease control strategies must be developed, and the determination of the genes and gene products that are associated with metabolic changes during host-pathogen interactions will provide the extensive insight that is needed to understand the host defense, disease and its control.
Plant-pathogen interactions were outlined in a four-phased ‘zigzag’ model by Jones and Dangl (2006). Pathogen recognition is a critical point in plant defense induction. The most important difference between resistant and susceptible plants is the timely perception of pathogenic signal molecules and the effective activation of the host defense mechanisms. According to the zigzag model, the first defense responses in plants, known as basal defense, act against all potential pathogens and are induced by the recognition of conserved pathogen/microbe-associated molecular patterns (PAMPs/MAMPs). The recognition of these molecules activates PAMP-triggered immunity (PTI), which induces the expression of defense response genes. In addition, antimicrobial compound production, reactive oxygen species (ROS) accumulation, callose deposition at the site of infection to strengthen the cell wall, the induction of signal molecules such as phytohormones (salicylic acid, jasmonates and ethylene) and the production of various PR proteins in neighboring cells accompany this activation (Jones and Dangl 2006; Pieterse et al. 2009). To counteract the host defenses, most pathogens, including rust pathogens, secrete an array of proteins that are known as effectors; they suppress the PTI and promote virulence. In fact, effectors have dual activities in the host as virulence promoters (virulence factors) and/or defense inducers (avirulence factors (Avr) or elicitors). As virulence factors, the effectors of the rust fungi move inside the plant cells via specialized infection structures known as haustoria and manipulate the host cell structure, metabolism and function to facilitate pathogenesis (Hogenhout et al. 2009).
To respond to this suppression and invasive attack, plants employ a second immune system, termed effector-triggered immunity (ETI), based on Avr recognition by disease resistance (R) proteins that are both intra- and inter-cellular receptors and specifically recognize the Avr either directly or indirectly. ETI leads to accelerated and amplified PTI responses and frequently results in the localized programmed cell death of infected plant cells, known as the hypersensitive response (HR) (Jones and Dangl 2006; Dixon et al. 2000). This response limits particularly biotrophic pathogen invasion due to the reliance of these pathogens on living host cells for nutrition and water (Glazebrook et al. 1997). In addition, the cells undergoing programmed cell death produce signals that activate the immune response termed systemic acquired resistance (SAR), which enhances disease resistance against a broad spectrum of pathogens with long-lasting effects in adjacent cells and in distant tissues (Durrant and Dong 2004). SAR activates the expression of pathogenesis-related (PR) genes and leads to the induction of extensive transcriptional reprogramming of defense-related genes (Jones and Dangl 2006). The same defense genes and a similar signaling network are used by both ETI and PTI but the activation and occurrence of these genes in ETI are stronger and faster than they are in PTI (Tao et al. 2003).
In addition to the most complex biological processes which are accompanied to defend/attack responses in plant-pathogen interactions, through co-evolution both plants and their pathogens have developed various elaborate mechanisms to survive and overcome pathogen attack or plant defense. Hence, to reveal the active host- and pathogen-derived proteins during the development of infection is a promising way of understanding of these complex interactions. Besides, this type of research has the potential to explore target proteins/genes such as effectors that can be used in disease management strategies. In recent years, rapidly developing proteomic technology has become a very important tool for providing direct insight into this extremely complex relationship and significantly contributes to understand how changes in gene expression becomes a cell response by completing the genomic and transcriptomic data.
In this study, proteomic analyses were conducted to determine the host and pathogen-derived proteins that contribute to compatible wheat-Pst interactions at different time points of the infection. We identified 102 host-derived and 64 pathogen-derived unique proteins and evaluated them within the context of the wheat-rust pathosystem. In addition all identified proteins were searched by bioinformatics-based algorithms to detect their subcellular localisation and signal peptide motifs to seek candidate effector proteins.
Material and methods
Plant material, Pst inoculation and sampling
A highly susceptible bread wheat (Triticum aestivum L.) cultivar, Seri82, to the fungal pathogen Puccinia striiformis f. sp. tritici was used in this study to form a compatible interaction. Plant seeds and pathogen urediniospores were provided by the Central Research Institute for Field Crops (Turkey). The Pst isolates were virulent to Yr2, Yr6, Yr7, Yr8, Yr9, Yr11, Yr12, Yr17, Yr18, Yr27 and YrA+ and avirulent to Yr1, Yr5, Yr10, Yr15, Yr24, YrSP and YrCv. Eighty seeds of Seri82 were planted in pots (7 cm in diameter) that were filled with sterile peat. Each pot contained 4 seeds and was incubated in a greenhouse at 20 °C with a 16/8 h day/night photoperiod (110 μE m−2 s−1 photon flux density). Thirteen days after germination, at the two-leaf stage, half of the seedlings were inoculated by spraying with a 10 mg/mL suspension of urediniospores in light mineral oil (Soltrol 170, ChemPoint, Limburg, Netherlands) by special inoculater. The remaining half were mock-inoculated (control) with an equivalent volume of spore-free mineral oil. Fifteen minutes after inoculation, the plants were transferred to dark dew chambers with a humidity of 95–100 % at 9 °C for 24 h. After this period, the plants were transferred to a greenhouse with a 15 °C dark cycle and 25 °C light cycle. The first leaves of the Pst- and mock-inoculated seedlings were harvested at 1, 2, 3 and 4 days post inoculation (dpi) and immediately placed in liquid nitrogen. These leaves were stored in a − 80 °C freezer until protein extraction. Three independent biological replicates were analyzed in this study.
Disease assessment
The compatible interactions were monitored by microscopy (Leica DMI6000B, Wetzlar, Germany) of the second leaves at 5 dpi. The hyphal growth of the Pst spores in the leaves was determined by lactophenol trypan blue staining. Leaf samples (~ 0.5 × 0.5 cm2) of infected and control samples put into 2.5 ml clearing solution A (acetic acid: ethanol, 1: 3, v/v) and shaken at low speed for overnight. Next day, solution A removed and 2 ml clearing solution B (acetic acid: ethanol: glycerol, 1: 5: 1, v/v/v) was added and shaken at low speed for at least 3 h. Solution B was removed and staining solution (0.01 % trypan blue in lactophenol) was added to each sample and they were shaken at low speed for overnight. Next day, the leaves were rinsed with sterilized 60 % glycerol, placed on the slides and monitored with microscope. In addition, the infection type (IT) was recorded using second leaves on a 0-to-9 scale (McNeal et al. 1971) at approximately 15 dpi.
Protein extraction
The protein extraction was performed using the combined protocols of Kim et al. (2001) and Rampitsch et al. (2006). Two grams of leaf samples were ground to a fine powder in liquid nitrogen using the Retsch MM301 system. The powder was homogenized in 20 ml ice-cold Mg/NP-40 extraction buffer containing 0.5 M Tris-HCl (Roche 122010) pH 8.3, 2 % (v/v) NP-40 (Sigma I7771), 20 mM MgCl2 (Sigma M8266), 2 % (v/v) beta-mercaptoethanol (AppliChem A1108), 1 mM PMSF (Sigma P7626) and 1 % (w/v) PVPP (Fluka 77627). After centrifugation at 12,000×g for 15 min at 4 °C, the proteins in the supernatant were fractionated with 15 % PEG 4000 (Sigma 95904). The samples were incubated on ice for 30 min and then centrifuged at 15,000×g for 10 min. The supernatant was precipitated by adding four volumes of cold acetone and was then kept at −20 °C overnight. After centrifugation at 12,000×g for 20 min at 4 °C, the pellet was washed 6 times with cold acetone. The pellets were dissolved in 2 ml solubilization buffer [7.5 M urea (Sigma U0631), 2.5 M thiourea (Sigma T7875), 12.5 % (v/v) glycerol (Sigma C3023), 62.5 mM Tris (Roche 122010), 2.5 % (w/v) 1-n-octylglucopranoside (Sigma 08001), 6.25 mM TCEP (Sigma C4706) and 1.25 mM protease inhibitor cocktail (Sigma P2714)]. The solubilized sample was sonicated five times for 5 s and centrifuged at 30,000×g for 30 min; then, the supernatant was centrifuged at 90,000×g for 1 h. The final protein content was determined using the Bradford Microassay procedure (Bio-Rad Laboratories) with BSA as a standard. The samples were stored in aliquots (500 μg/500 μl) at −80 °C.
First-dimension fractionation by PF2D
The ProteomeLab™ PF2D system (Beckman-Coulter, Fullerton, CA) is a 2D-liquid chromatography system for the two-dimensional separation of protein mixtures. The proteins were separated in the first dimension according to their isoelectric point (pI) using the chromatofocusing method.
The first-dimension separation was performed at room temperature (24 ± 1 °C) with two buffers (Start Buffer pH 8.5 and Eluent Buffer pH 4.0). These buffers were freshly prepared according to Barré and Solioz (2006). The high-performance chromatofocusing (HPCF) column was maintained according to the manufacturer’s instructions (Eprogen A51680). The column was washed with water at a 0.2 ml/min flow rate for 45 min, and the Start Buffer began to flow during the 130th min at a 0.2 ml/min flow rate to equilibrate the column. At the same time, the extracted protein samples were desalted on a PD-10 SephadexTM G-25 gel filtration column and eluted with 3.5 ml chromatofocusing Start Buffer. The protein quantification was performed using the micro BCA Protein Assay Kit (Sigma QPBCA). After column equilibration, 3 mg desalted protein sample were loaded onto the column using a manual injector. In the first dimension, the proteins bound to a strong anion exchanger, and the pH began to decrease from 8.5 to 4.0 after 60 min. The proteins were eluted with a continuous decreasing gradient, and the fractions were collected at 0.3-pH intervals in a 96-deep-well plate.
Second-dimension fractionation by PF2D
The PF2D second-dimension separation utilizes reverse phase high performance liquid chromatography (HPLC) fractionation. The proteins that were eluted during the pH gradient in the first dimension were separated in a second dimension by their hydrophobic properties. Two solvents were used for the hydrophobicity gradient: 0.1 % TFA (v/v) in HPLC water (Solvent A) and 0.08 % TFA (v/v) in acetonitrile (ACN) (Solvent B). The separation was performed at 50 °C with a 0.75 ml/min flow rate, and the protein absorbance was detected using a UV2 detector at 214 nm for each fraction. A high-performance reversed phase (HPRP) (Eprogen A51689) column equilibration was achieved with Solvent B for 10 min, followed by Solvent A for 5 min for each injection. From the selected first-dimension fractions, 0.2 ml sample was injected into second-dimension module and linear gradient of 0–100 % Solvent B for 25 min is run. The proteins were collected at 0.75-min intervals, starting at 5 min and ending at 25 min. At the end of each 2nd- dimension run, Solvent B continued for 5 min and was followed by re-equilibration with 100 % Solvent A for 10 min.
The 32 Karat™ Software was used for data processing and calculating the peak areas and heights which represent abundance level of the protein. The protein profile for each sample was generated as a 2D-Map using the ProteoVue software. The Pst-inoculated and mock-inoculated protein profiles from three biological replicates were compared, and a peak-to-peak analysis of their chromatograms was performed using the DeltaVue software. In our previous study (unpublished data), three technical replicates were performed, and the area of 200 peaks was statistically analyzed using a t-test to calculate the minimum fold-change values for selecting the differentially expressed proteins. According to this analysis, the peaks with protein abundance levels that were more than 2-fold different between the Pst- and mock-inoculated samples were considered significant. Area of these identical peaks from each time point belonging to three biological replicates were statistically analysed by two-tailed Student’s t-test with SPSS software version 22.0. The peaks that have p < 0.05 were selected for identification.
Third-dimension separation by SDS-PAGE
After 2D protein fractionation, some selected fractions included additional proteins and required further separation. These fractions were dried and resuspended by adding 10 μL 50 mM NH4HCO3 (Fluka 09830); they were then analyzed by 12 % 1D SDS-PAGE as the third dimension. The protein bands were monitored by staining with Oriole™ Fluorescent Gel Stain (Bio-Rad 161–0497). The differentially expressed proteins bands of interest were excised and subjected to tryptic in-gel digestion.
Tryptic digestion of proteins
The disulfide bonds of proteins in the selected second-dimension fractions were reduced by adding 1 μL 100 mM Dithiothreiotol (DTT) (Sigma 43,815) and incubating at 60 °C for 1 h. The reduced cysteine side chains were alkylated by the addition of 1 μL 200 mM Iodoacetamide (IAA) (Sigma I1149) and incubated in the dark at room temperature for 30 min. The proteolytical digestion was performed by the addition of 0.2 μg proteomic-grade trypsin (Sigma T6567) in 50 mM NH4HCO3 and incubating at 37 °C overnight. In addition, the excised gel bands from the third-dimension separation were washed sequentially with 50 mM NH4HCO3 buffer and ACN, and the proteins were digested according to Wilm et al. (1996). The peptides were purified using a C18 ZipTip® (Millipore) according to the manufacturer’s recommendation. The eluates were dried under vacuum and resuspended in 5 μL HPLC-grade water with 0.1 % formic acid (Sigma 94318) and 50 fmol calibrant (ADH1_YEAST- Waters MassPrep Enolase Digestion Standard, 186002325) for mass spectrometric analysis.
Protein identification by nanoLC-ESI-MS/MS
Two microliters of peptide samples were loaded onto the system [nanoACQUITY ultra pressure liquid chromatography (UPLC) and SYNAPT high definition mass spectrometer] using a NanoLockSpray ion source. Prior to the injection, the columns were equilibrated with 97 % mobile phase A (water with 0.1 % FA) and 3 % mobile phase B (ACN containing 0.1 % FA). The column temperature was set to 35 °C. First, the peptides were trapped on a nanoACQUITY UPLC Symmetry C18 Trap column (5 μm particle size, 180 μm i.d. × 20 mm length) at a 5 μl/min flow rate for 5 min. The peptides were separated from the trap column via gradient elution onto an analytical column (nanoACQUITY UPLC BEH C18 Column, 1.7 μm particle size, 75 μm i.d. × 250 mm length) at a 300 nl/min flow rate, with a linear gradient from 5 to 40 % ACN over 90 min. The data-independent acquisition mode (MSE) was carried out by operating the instrument in positive ion V mode and applying the MS and MS/MS functions at 1.5-s intervals, with 6 V low-energy and 15–40 V high-energy collisions to collect the peptide mass-to-charge ratio (m/z) and the product ion information to deduce the amino acid sequence. To correct for the mass drift, the internal mass calibrant Glu-fibrinopeptide was infused every 45 s through the NanoLockSpray ion source at a 300 nl/min flow rate. The peptide signal data between 50 and 1600 m/z values were collected.
Tandem mass spectra extraction, charge state deconvolution and deisotoping steps were processed using the ProteinLynx Global Server (PLGS) V2.4 (Waters Corp, Milford, MA) and searched with the IDENTITYE algorithm against either the wheat database or the Broad Institute Puccinia Database (http://www.broadinstitute.org/). IdentityE was set to search null, assuming the digestion enzyme trypsin, and was searched with a fragment ion mass tolerance of 0.028 Da and a parent ion tolerance of 0.011 Da. The Apex3D data preparation parameters were set to a 0.2 min chromatographic peak width, a 10.000 MS TOF resolution, 150 counts for the low energy threshold, 50 counts for the elevated energy threshold and 1200 counts for the intensity threshold. The databank search query was set to a minimum of three fragment ion matches per peptide, a minimum of seven fragment ion matches per protein, a minimum of one peptide match per protein and 1 missed cleavage. The carbamidomethyl-cysteine-fixed modification and acetyl N-TERM, the deamination of asparagine and glutamine as well as the oxidation of methionine variable modifications were set. In addition, the amino acid sequences of some peptides that did not match with any known protein were searched in the NCBI databases using BLASTp.
Bioinformatic analyses
Bioinformatic analyses were performed for detection of potential signal-peptide signature and prediction of cellular localization. Firstly, proteins were analysed by WoLF PSORT (Horton et al. 2007) for domains and motifs that would indicate their targeting and/or cellular localisation. For this analysis, WoLF PSORT was run with the “plant database” and “fungi database” to analyse the host-derived proteins and pathogen-derived proteins, respectively. Signal peptide and N-terminal signal peptide prediction were performed with SignalP (v4.1) (http://www.cbs.dtu.dk/services/SignalP/) and TargetP (v1.1) (http://www.cbs.dtu.dk/services/TargetP/), respectively, using the default settings for eukaryotes. While cut off value is defined as 0.05 in TargetP, sensitive mode was choosen in SignalP.
Results and discussion
Assessment of wheat-Pst interaction
The compatible wheat-Pst interactions were verified by microscopic and macroscopic evaluations. At 5 dpi, developing infection germtubes or hyphaes from urediniospores on the Pst-inoculated susceptible Seri82 leaves were monitored by lactophenol-trypan blue staining under a microscope (Fig. 1a). At 15 dpi, heavy sporulation was observed on the infected leaves (infection type 9, a highly susceptible reaction) (Fig. 1b). All of the mock-inoculated plants were free from any disease symptom during the 15-day observation period (Fig. 1c and d). These observations indicate that successful compatible interactions were developed and leaves of the heavily infected plants which produced the highest IT (8–9) and control plants were used for proteomic analyses.
Microscopic and macroscopic visualisation of compatible interaction following Pst inoculation on a susceptible (Seri82) wheat genotype. Whole leaves at 15 dpi were used for macroscopic visualisation. Microscopic images were taken with a Hitachi 3-CCD HV-D20P color camera. a Pst-inoculated Seri82 at 5 dpi (Leica DMI6000B, 40× magnification); b Pst-inoculated Seri82 at 15 dpi; c Mock-inoculated Seri82 at 15 dpi; d Mock-inoculated Seri82 at 5 dpi (Leica DMI6000B, 40× magnification
Generation and comparison of protein profiles
The protein extracts of the Pst-inoculated and mock-inoculated leaf samples that belong to three independent biological replicates at 1, 2, 3 and 4 dpi were analyzed to determine the differentially expressed proteins during the compatible interactions between wheat and Pst. For each sample, 3.0 mg protein extract was injected into the system and separated two-dimensionally by PF2D, which permits the two-dimensional fractionation of the intact proteins. In general, the recovered proteins in the first dimension were concentrated in two regions during the pH gradient as follows: 21 % of the proteins were in pH ≥8.5, and 76 % were between pH 7.0 and 4.0 (Fig. 2). At the end of the pH gradient, non-protein peaks were observed, while a higher NaCl peak was observed in the 29–31 fractions due to the washing buffer. Therefore, the first 23 fractions were injected into the HPRP module for second-dimension separation. The reproducibility of the fractionation step completely depends on the reproducibility of the pH gradients. Freshly prepared Start and Eluent Buffer in the same lot were used for the separation of all of the samples at one time point. Following the two-dimensional separation, to view and compare the proteome profiles of the mock- and Pst-inoculated samples, virtual 2D gel images were formed by combining the first-dimension pH data and the second-dimension UV absorbance (214 nm) data using the ProteoVue tool (Fig. 3).
High reproducibility of two dimensional seperation of leaf protein extracts by ProteomeLab PF2D. Panels a and b show also a high degree of technical reproducibility. a Representative comparison of protein UV (214 nm) chromatograms of mock- (solidline) and Pst-inoculated (dashedline) tissue after injection of the identical 1st dimension fractions (Seri82_3 dpi/19. fraction) of the each biological replicates. b Comparison of same fraction chromatograms of the three independent biological replicates both mock- and Pst-inoculated groups
High reproducibility is one of the most important requirements of proteomic studies. The reproducibility of the PF2D system was examined by comparing the virtual 2D maps and second-dimension UV chromatograms of the mock- and Pst-inoculated samples between three biological replicates at each time points (Fig. 3). As shown in Fig. 3, the precise alignment of peaks among the replicate runs clearly indicates that a high-resolution and reproducible protein separation was obtained in this study. In addition to reproducibility, the identical protein peaks were analyzed using the two-tailed Student’s t-test with SPSS software version 22.0 among three biological replicates. For this, only the peaks which their abundance difference greater than two fold between Pst- inoculated and control samples were statistically evaluated.
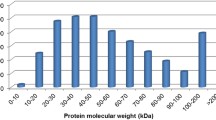
To determine the differentially expressed proteins during the compatible interactions, 2D maps of the mock- and Pst-inoculated leaf samples were compared, and quantitative and qualitative peak-to-peak analyses were performed using the paired peak function in the DeltaVue mapping tool (Fig. 4). On average, more than 700 unique peaks were reproducibly compared in the protein extracts from each time point, and many differentially expressed proteins were detected (Fig. 5). The peaks that were at least two-fold different from controls and p < 0.05 were selected for identification. Some of the selected fractions that included additional proteins and required further separation were evaluated after additional separation by 1D SDS-PAGE (Supplementary Fig. S1). The selected fractions from the PF2D and the protein bands from the PAGE were identified by nanoLC-ESI-MS/MS.
Functional classification and their ratio of the identified proteins which were differentially expressed in susceptible (Seri82) wheat leaves following Pst inoculation. Proteins were classified into seven groups based on their roles in diverse biological processes. The table shows compared protein peak numbers and identified proteins numbers for each time point
Protein identification
Many proteins whose abundance changed during the compatible interactions between wheat and Pst were identified. 212 proteins (35, 29, 91 and 57 proteins at 1, 2, 3 and 4 dpi, respectively) were identified as differentially expressed in the infected leaf tissue compared with the mock-inoculated control. In total 126 proteins include unique peptide and 166 proteins were unique. Sixty two percent of total had a host origin (Table 1, Table S1), and 38 % were pathogen-derived (Table 2, Table S3). Both the identified plant and fungal proteins were categorized into different functional groups based on their molecular functions, which were provided by the databases and the available literature. The relative representation of each group and the percentages of the identified plant proteins showed variations among the different time points (Fig. 5). In general, the most abundant plant proteins at all of the time points were involved in the defense response, stress, photosynthesis and metabolism. The remaining plant proteins were related to signal transduction, gene expression and electron transport, and often lower than 10 %. Another significant portion of the identified proteins (average 30 % at each time point) were pathogen-derived fungal proteins. These proteins were also classified into different functional groups, including attack, signal transduction, gene expression, metabolism and structure. Most of the fungal proteins identified during the late stages of the infection (3 and 4 dpi) did not show homology with amino acid sequences of any known proteins and matched only hypothetical proteins from the studied wheat pathogenic fungi Phaeosphaeria nodorum, Puccinia graminis f. sp. tritici and Puccinia triticina. While the number of proteins in the attack functional group, which included proteins that were important for the penetration and invasion of Pst and the suppression of plant defense, was greater than 20 % in the first 2 days after inoculation, this number decreased to 3 % at 3 and 4 dpi. However, the number of hypothetical proteins increased significantly (>60 %) at the same time points. This results was considered as a promising opportunity to detect unknown pathogenicity factors such as effectors. Hence all the identified proteins were analyzed using WoLF PSORT and a combination of SignalP and TargetP. This analyses revealed that nine fungal proteins have a N-terminal signal peptide and two proteins have a mitochondrial targeting peptide (Table 2). Seven of them (PGTG_07231T0, CBH50687.1, ABA42174.1, EFQ63896.1, EFQ62190.1, EFQ64083.1, ZP_07774948.1) were predicted as extracellular proteins. Among these, three hypothetical proteins have features of the known effector proteins such as small peptide (˂300 aa), extracellular localization, and high proportion of cysteine amino acids. It is further discussed in the “Pathogen-derived Proteins” section. Detailed data of wheat and fungal proteins for all of the time points are presented as supplementary data (Tables S1, S2, S3 and S4).
In this study, we identified numerous proteins (Table S1 and S3) during compatible interaction between wheat and stripe rust. Therefore in the following sections, only proteins which have potential roles in host defense and pathogen attack are discussed within the context of the cereal-fungus pathosystem.
Host-derived proteins
The molecular research on the host-pathogen relationship revealed that similar defense responses are used by both resistant and susceptible cultivars, but that their activation and occurrence in compatible interactions seem weaker and occur later. This situation is reviewed as differences between susceptibility and resistance are associated with differences in the timing and magnitude of changes rather than with the expression of different sets of genes. Our results which are represented below also support this hypothesis.
In the current study, identified host-derived proteins were classified in the 7 functional groups included defense, stress responsive, signal transduction, gene expression, photosynthesis, electron transport and metabolism. Among these, the most abundant plant proteins at all of the time points were involved in the defense- and stress-related proteins (Fig. 5). Among the defense class the antioxidant and detoxification proteins were the major group included superoxide dismutases (SODs), glutathione S transferase (GST), catalase (CAT), peroxidases (PX), peroxiredoxins (PRX), ascorbate peroxidase (APX) and dehydroascorbate reductase (DHAR) (Table 1).
Oxidative burst is one of the earliest plant defense responses and is characterized by the rapid production and accumulation of reactive oxygen species (ROS) following exposure to biotic and abiotic stress. ROS are toxic to both plant and pathogen cells and also play important roles as signal molecules for the activation of the defense responses in host (Mendoza 2011). In addition, ROS are cell wall reinforcement materials and diffusible signals for programmed cell death (PCD) induction (Mellersh et al. 2002). However, pathogens also benefit from ROS due to the development of specialized penetration structures and successful infection (Nanda et al. 2010). Tenberge et al. (2002) reported a cytochemical study that showed O2· − accumulation around the hyphal tips and H2O2 generation around the penetrated cell during the infection process of B. cinerea. Only SOD enzymes (cytosolic and chloroplastic Cu/Zn-SOD, mitochondrial SOD and Mn-SOD) were identified as antioxidants at 1 dpi in the susceptible cv. Seri82; however, in the following stages (2, 3 and 4 dpi), other antioxidant proteins (CAT, PX, PRX, GT, GST, GPX, APX, DHAR and their isoforms) in addition to SOD were identified. All of these proteins were over-represented or induced (only expressed in the infected samples). It has been suggested that the infected wheat cells were under the heavy bombardment of ROS that may have been produced by Pst for attack and by the plant for defense. Similarly, Garcı́a-Limones et al. (2002)reported that the APX, CAT, SOD and GPX enzymes were significantly induced during disease development in chickpea caused by Fusarium oxysporum f. sp. ciceri. In contrast to the compatible interaction, many antioxidant enzymes (CAT, PX, PRX, GST and their isoforms), but not SOD, were identified in incompatible wheat-Pst interaction as up-regulated defense-related proteins (Maytalman et al. 2013). In many incompatible interactions, the overexpression of these enzymes in the host activates plant resistance and protects the host cells by regulating the peroxide concentrations outside the infected tissues and thereby sparing the uninfected cells (Nanda et al. 2010; Rizhsky et al. 2002).
The second major group of proteins of the defense class was pathogenesis-related proteins (PR) which are the most important members of the plant defense (Table 1). Most of the PR proteins have an antimicrobial, antifungal or antiviral effect, and the accumulation of these proteins has been reported in many plant species during the early stages following pathogen attack (Van Loon 2006). The expression profiles of PR genes differ from species to species and in response to different pathogens. In general, the overexpression of the PRs is considered a direct sign of the resistance response; however, in some species, PRs are also induced during disease development in response to different pathogens (Hou et al. 2012; Elvira et al. 2008; Hammond-Kosack and Jones 1996). For instance, Xin et al. (2012) found that most PR genes were induced to a higher level in a susceptible genotype than in a resistant genotype in a comparative transcriptomic study between compatible and incompatible interactions between wheat and powdery mildew. They interpreted these results that PRs are related to the severity of symptom expression rather than to resistance. Similarly, in our study, many PR proteins were identified only in the infected leaves during compatible interaction. PR1 species (PR1.1, PR1.2, PR1.3) and PR4 were identified as direct PR proteins during the late stage (3 dpi). In addition, some defense-related proteins that were considered PR protein families in different plant species based on their antifungal activity against specific pathogens and possessed enzymatic or inhibitory activities (Bertini et al., 2009), such as chitinases (PR3, PR8, and PR11), proteinase inhibitors (PR6), sulfur-rich thionin-like proteins (PR13) and lipid transfer proteins (PR14), were identified at 2 and 3 dpi.
The PR1 proteins, known as hallmarks of defense pathways, are mostly overexpressed upon infection. However, in diverse plant species, antifungal effects of PR1 proteins were shown in vitro, and recent studies suggest that the biological functions of the PR1 proteins are still obscure. There are examples, similar to our results, which the accumulation of PR1 proteins was induced during both compatible and incompatible interactions (Lu et al. 2011). This observation was evaluated as the expression levels and the timing of regulation PR1 family proteins may be critical in determining the outcome of the host defense responses. PR4 proteins possess antifungal activity against several pathogenic fungi and block hyphal growth and spore germination by inhibiting the pathogenic translation process via their ribonuclease activity (Caporale et al. 2004). Bertini et al. (2003) reported that PR4 proteins are also specifically induced in wheat following fungal infection and SAR activation. In addition, PR4 proteins play important roles in the HR, which is characteristic of the resistance response in wheat against yellow rust (Wang et al. 2010). However, similar to our results, Bozkurt et al. (2010) showed that the expression levels of the PR2 and PR4 transcripts increased in susceptible wheat cultivar at 24 and 48 h after Puccinia striiformis inoculation. Vaghefi et al. (2013) also reported that PR4 gene expression was induced upon fungal infection in both resistant and susceptible cultivars of lentil and that the magnitude of PR4 expression was higher in the susceptible cultivar.
Chitin is a major component of fungal cell walls and is recognized by plants as a general PAMP (Boller 1995). Therefore, fungal attack triggers the accumulation of chitinases, which have the direct effect of limiting fungal invasion by degrading the fungal cell wall. Moreover, the produced chitin fragments function as elicitors of numerous downstream defense response genes (Eckardt 2008). In this study, chitinase and endochitinase, which are classified in the PR protein families as PR3, PR8 and PR11, were identified among other defense-related proteins in the compatible interaction at 3 and 4 dpi. Similar results were also obtained by Legay et al. (2011) in their suppression subtractive hybridization (SSH) study that identified of genes expressed during the compatible interaction of grapevine with Plasmopara viticola. We suggest that this is an expected result because the susceptible host consistently faces increasing pathogen invasion during infection.
Proteolysis and protease inhibition are important aspects of pathogen infection and host defense. Proteases may be used in different processes, such as enabling both the penetration of the cell wall and the destruction of defense-related proteins or cell wall proteins during colonization by fungal phytopathogens (Scholtz 2013). In addition, some virulence factors of pathogenic species are extracellular proteases, such as cysteine, serine, aspartic and metalloproteases (Alkan et al. 2013). Therefore, protease inhibitors are crucial components of the plant defense response by counteracting exogenous proteases secreted by pathogens (Yang and Yeh 2005). In this study, 2 members of the cysteine protease inhibitor superfamily (cysteine proteinase inhibitor and cystatin WC1) and 2 members of the serine protease inhibitor family (Wali3 and Wali5) were up-represented at 3 dpi. The over-representation of serine protease inhibitors in wheat defense against pathogens has been shown in a number of studies. The induction of wali5 and WRSI5 expression was shown in a cDNA library that was derived from a Puccinia triticina (Pt)-resistant wheat cultivar by Manickavelu et al. (2010). In another study, Scholtz (2013) reported that the expression levels of Wali5 were similar in the resistant and susceptible wheat genotypes during Pt and Pst infection. However, the current study and our previous qRT-PCR studies (unpublished) have shown the significant induction of Wali5 expression in the compatible wheat-Pst interaction.
In several plant-pathogen studies, LTPs were closely related to plant defense mechanisms and were consequently classified as pathogenesis-related proteins under the denomination PR14 (Van Loon and Van Strien 1999). Maldonado et al. (2002) reported that some LTPs contain signal peptides for defense activation and exhibit antimicrobial activity. Subsequent studies introduced the role of LTPs, especially in plant fungal resistance. In one of these, it was reported that LTPs lead to fungal cell death by inserting themselves into fungal cell membranes and forming a pore which causes efflux of intracellular ions (Selitrennikoff 2001). Besides, in rice, LTPs inhibited the germination of Pyricularia oryzae spores (Ge et al. 2003) and affected the fungal appressoria formation and the penetration of Glomus mosseae (Blilou et al. 2000); in wheat, LTP1 expression was increased in the first 12 h following inoculation and reduced the penetration efficiency of Blumeria graminis f. sp. tritici in a susceptible cultivar by approximately 28.3 % (Li et al. 2006). However in our study, LTP1 (at 3 dpi) and LTP2 (at 2 and 3 dpi) were up-represented in the late stage of the infection and did not block the pathogen invasion.
Flavonoids are ubiquitous secondary metabolites and include most antimicrobial and antifungal phytoalexins, important signaling molecules and mediators that are associated with plant defense. Therefore, several enzymes that are involved in flavonoid biosynthetic pathways are up-regulated during plant-pathogen interactions. However, increased expression levels of some were also reported in susceptible genotypes following pathogen attack (Fang et al. 2013; Xin et al. 2012). In this study, flavanone 3-hydroxylase (F3H), which is one of the key enzymes of the flavonoid pathway, was identified in the susceptible cv. Seri82 at 1 dpi as potentially another important defense response element in wheat against Pst.
Pathogen recognition is an important critical point of plant defense activation. Several types of cell-surface receptors in plants perceive diverse pathogen- and environment-derived signals. In wheat, receptor-like kinases (RLKs), including RLK-R3 and LRK10, constitute a large family of these receptors (Walker 1994). In many studies, plant RLKs were mainly involved in the defense response to biotic and abiotic stresses (Shiu and Bleecker 2001; Zhou et al. 2007). Similarly, in our study, the resistance-related receptor-like kinases (RLK-R3) and rust-resistance kinase Lr10 (LRK10) were identified as defense-responsive proteins in susceptible cv. at 2 dpi. The molecular and functional properties of wheat RLKs, TaRLK-R1, TaRLK-R2 and TaRLK-R3 under stress conditions were investigated by Zhou et al. (2007). These authors reported that TaRLK-R3, at least, is functionally involved in both the resistance to stripe rust infection and the response to abiotic stresses in wheat plants. These authors also showed that TaRLK-R1, TaRLK-R2 and TaRLK-R3 are positive contributors to the wheat HR against stripe rust fungus. The most studied member of the wheat leaf rust kinase (WLRK) gene family is LRK10, which is genetically associated with the Lr10 locus and confers resistance to leaf rust fungus (Feuillet et al. 2003). On the other hand RLKs are involved in not only defense response as they are also responsible for growth and development of plants (Shiu and Bleecker 2001; De Smet et al. 2009). Some RLKs possess roles in immunity and growth, such as brassinosteroid receptors LRR-RLK-BAK1 and bacterial PAMP flagellin. Flagellin perception changes endogen hormone levels in plants and thus develop disease resistance or succeptibility, for instance flagellin inhibits auxin signaling and activates resistance mechanisms against biotrophic pathogens (Navarro et al. 2006) whereas perception of exogenous protein such as pathogen effectors trigger giberellic acid and auxin production as virulence strategies (Grant and Jones 2009).
Pathogen-derived proteins
Sixty four pathogen-derived fungal proteins, which averaged 30 % of the total proteins at each time point except 2 dpi, constituted a significant portion of the identified proteins in the compatible interaction. Thirty of them were classified in the five functional groups included attack, signal transduction, gene expression, metabolism, and structure. The remaining large group (34 of total fungal proteins and ~70 % of the identified proteins at 3 and 4 dpi) were hypothetical proteins of unknown function (Fig. 5 and Table 2). These results suggest that these late-stage hypothetical proteins could be related to the virulence of pathogens and nutrient uptake from host cells, to the production of infection structures and to invasion.
Biotrophic plant pathogens such as rust pathogens secrete an array of proteins, known as effectors (Hogenhout et al. 2009). They are virulence tools of phytopathogens to suppress host basal defense and facilitate infection. Effectors are secreted into the plant apoplast or delivered into host cells by infectious structures such as appresoria and haustoria to play an important role in disease development (Koeck et al. 2011). Therefore the identification and characterisation of effectors is very important to develop effective and sustainable strategies for management of the phytopathogens. Hence the secreted proteins during plant-pathogen interaction is one of the significant areas of interest in plant proteomics (Agrawal et al. 2010). However, recovery of the secreted fungal proteins from infected plant tissues, very low abundance of pathogen-derived proteins in comparison to the host-derived, cytoplasmic contamination and limited secretome database are difficulties in effector investigations.
Effector proteins must first be secreted, and the synthesis and secretion of such proteins appears to be one of the primary functions of haustoria. This is generally assumed to be accomplished through the action of N-terminal signal or transit peptides, which are cleaved off to create the mature effector proteins. Effector protein candidates can be detected using a bioinformatics approach, however, only known sequences can be used since no universal signalling mechanism has been found in rusts (Rampitsch et al. 2015). In the current study, we analysed all fungal proteins bioinfomatically to detect effector candidates. This analysis revealed that nine fungal proteins have N-terminal signal peptides and seven (PGTG_07231T0, CBH50687.1, ABA42174.1, EFQ63896.1, EFQ62190.1, EFQ64083.1, ZP_07774948.1) were predicted as extracellular proteins. All of these have a predicted signal peptide (Table S4), however just three uncharacterised proteins, PGTG_11681T0, PGTG_07231T0 and CBH50687.1, have the known effector protein features such as small size (˂300 aa), extracellular localization, and cysteine-rich composition. Their cysteine contents were 0.4 % for PGTG_07231T0, 3 % for CBH50687.1 and 4.6 % for PGTG_11681T0. Although these proteins have been identified as candidate effectors, the best candidate is PGTG_11681T0 according to its cysteine content and short peptide length. However, the subcellular localization prediction of PGTG_11681T0 is to the nucleus and this suggests that it may be a nuclear-localized effector. Recent studies showed that nuclear-localized effectors target the host nuclei through nuclear pores and modulate host transcription to suppress defense by interacting with the mediator complex that controls interactions between transcriptional regulators and RNA polymerase (Rovenich et al. 2014). In one of these, it was reported that HaRxL44, a nuclear-localized effector of downy mildew pathogen of Arabidopsis, interacted with MED19a (mediator complex subunit) and reduced SA-regulated gene expression (Caillaud et al., 2013).
During plant-pathogen co-evolution, plants and pathogens each developed many strategies against the other to survive. The pathogenic fungi evolved to generate a successful infection by developing bidirectional evolutionary methods: while these fungi developed methods to perceive, penetrate and invade the host and infection structures for nutrient uptake, they also developed approaches to escape from host recognition, suppress the host defense and detoxify the phytotoxins.
In this study, two fungal proteins that were identified in the susceptible cv. Seri82 at 1 dpi showed a high degree of similarity with methyl-accepting chemotaxis proteins (MCPs) and the flagellar protein FliO/FliZ of Pseudomonas fluorescens. Most microorganisms use chemical sensing, to find nutrients and escape from harmful compounds. MCPs are well known chemoreceptors in Bacteria and Archaea, and their coding genes have been identified in fungi and are highly conserved between the studied species (Blanco-Ulate et al. 2013). The importance of MCPs in the disease cycle, particularly in recognizing the host and initiating the infection, has been reported for several plant-microbe interaction including phytopathogenic fungal zoospores. It is now clear that fungal spores use physical or chemical signals from the plant surface to trigger germination and differentiation for the successful infection of the host (Hua et al. 2008). However MCP was not identified in rust fungi before sequencing Puccinia graminis f. sp. tritici genome that it was annotated as unusual family domain in SUPERFAMILY database.
Following the perception of host plant signals, many fungal pathogens activate an orderly sequence of morphological differentiation, including spore germination, germ tube elongation, appressorium formation and penetration hyphal development. Many studies of plant-fungus interactions have demonstrated that adhesion is essential for the development of these infection structures for several pathogens (Vidhyasekaran 2008). FliO is a short protein that is found in flagellar biosynthesis operons and is an essential component of the flagellum-specific protein export (Tomich et al. 2002). Although the molecular function of this gene is unknown, in some bacteria components of the flagellum may act as adhesions and play roles in colonization through direct interactions with host ligands (Yao et al. 1994). In addition, flagellum and flagellum-mediated motility contribute to the virulence of a number of pathogenic bacteria, and mutations in flagellar biogenesis genes attenuate the virulence of several human pathogens, including Pseudomonas aeruginosa, Proteus mirabilis, Campylobacter jejuni and Helicobacter pylori (Tomich et al. 2002). This protein is also a well known PAMP for bacterial pathogens, but in the absence of flagellar this was not identified as a PAMP/effector for fungal pathogens. However in our study, flageller protein, FliO/FliZ was detected and predicted as an effector candidate and also classified as an attack protein. The results of the Takakura et al. (2008) support our finding. In this study, they showed that the expression of a bacterial flagellin gene in transgenic rice triggers immune responses and enhances disease resistance to fungal infection, rice blast, caused by Magnaporthe grisea.
The suppression of the host defense is another crucial strategy that biotrophic pathogens use to establish the infection and induce the feeding site without host detection/defense. Isochorismatase hydrolase, which has been well characterized in pathogenic organisms (Caruthers et al. 2005), was considered by El-Bebany et al. (2010) to be a potential plant-defense suppressor that was produced by a highly aggressive isolate. In plants, the endogenous signal molecule SA is synthesized via the phenylpropanoid and isochorismate pathways from the precursor isochorismate. Isochorismatase hydrolase competes directly with the enzyme (currently unknown in plants) that is responsible for converting isochorismate into SA and it converts isochorismate, in the presence of water, to 2,3-dihydroxybenzoate and pyruvate and inhibits salicylic acid formation. It may be worth speculating that isochorismatases that are secreted by fungi could reduce SA accumulation in response to pathogen attack and thus inhibit plant defense responses (El-Bebany et al. 2010). Similar to previous plant-pathogen interaction studies, we identified isochorismatase hydrolase as a differentially expressed fungal protein in the susceptible cv. Seri82 at 3 and 4 dpi.
Most of the identified fungal proteins (~70 %) at 3 and 4 dpi were hypothetical. These results suggest that these late-stage hypothetical proteins could be related to nutrient uptake from host cells, to the production of infection structures, and to invasion.
Conclusion
In this study, proteome analyses were conducted to determine the differentially expressed host- and pathogen-derived proteomes during a compatible interaction between a susceptible wheat cultivar and Pst. The potential roles of the identified host defense-related proteins and pathogen-derived attack proteins are discussed within the context of the cereal-fungus pathosystem. Collectively, our results support the hypothesis that similar defense responses are used by both resistant and susceptible cultivars but that their activation and occurrence in compatible interactions seem weaker and occur later. In addition, three fungal proteins have been tentatively identified as candidate effectors. An addition protein similar to components of bacterial flagella is predicted as another effector candidate. Although this protein is a PAMP for bacterial pathogens, this needs to be examined for rust pathogens.
References
Agrawal, G. K., Jwa, N. S., Lebrun, M. H., Job, D., & Rakwal, R. (2010). Plant secretome: unlocking secrets of the secreted proteins. Proteomics, 10(4), 799–827.
Alkan, N., Espeso, E. A., & Prusky, D. (2013). Virulence regulation of phytopathogenic fungi by pH. Antioxidants & Redox Signaling, 19, 1012–1025.
Barré, O., & Solioz, M. (2006). Improved protocol for chromatofocusing on the ProteomeLab PF2D. Proteomics, 6, 5096–5098.
Bertini, L., Caporale, C., Testa, M., Proietti, S., & Caruso, C. (2009). Structural basis of the antifungal activity of wheat PR4 proteins. FEBS Letters, 583, 2865–2871.
Bertini, L., Leonardi, L., Caporale, C., Tucci, M., Cascone, N., Di Berardino, I., Buonocore, V., & Caruso, C. (2003). Pathogen-responsive wheat PR4 genes are induced by activators of systemic acquired resistance and wounding. Plant Science, 164, 1067–1078.
Blanco-Ulate, B., Rolshausen, P., & Cantu, D. (2013). Draft genome sequence of Neofusicoccum parvum isolate UCR-NP2, a fungal vascular pathogen associated with grapevine cankers. Genome Announc, 1(3), e00339–e00313.
Blilou, I., Ocampo, J. A., & García-Garrido, J. M. (2000). Induction of Ltp (lipid transfer protein) and Pal (phenylalanine ammonia-lyase) gene expression in rice roots colonized by the arbuscular mycorrhizal fungus Glomus mosseae. Journal of Experimental Botany, 51, 1969–1977.
Boller, T. (1995). Chemoperception of microbial signals in plant cells. Annual Review of Plant Physiology and Plant Molecular Biology, 46, 189–214.
Bozkurt, T. O., McGrann, G. R. D., MacCormack, R., Boyd, L. A., & Akkaya, M. S. (2010). Cellular and transcriptional responses of wheat during compatible and incompatible race-specific interactions with Puccinia striiformis f. sp. tritici. Molecular Plant Pathology, 11, 625–640.
Caillaud, M.-C., Asai, S., Rallapalli, G., Piquerez, S., Fabro, G., et al. (2013). A downy mildew effector attenuates salicylic acid–triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biology, 11(12), e1001732.
Caporale, C., Di Berardino, I., Leonardi, L., Bertini, L., Cascone, A., Buonocore, V., & Caruso, C. (2004). Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Letters, 575, 71–76.
Caruthers, J., Zucker, F., Worthey, E., Myler, P. J., Buckner, F., Van Voorhuis, W., Mehlin, C., Boni, E., Feist, T., Luft, J., et al. (2005). Crystal structures and proposed structural/functional classification of three protozoan proteins from the isochorismatase superfamily. Protein Science, 14, 2887–2894.
De Smet, I., Voss, U., Jürgens, G., & Beeckman, T. (2009). Receptor-like kinases shape the plant. Nature Cell Biology, 11, 1166–1173.
Dixon, M. S., Golstein, C., Thomas, C. M., van der Biezen, E. A., & Jones, J. D. G. (2000). Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2. Proceedings of the National Academy of Sciences of the United States of America, 97(16), 8807–8814.
Durrant, W. E., & Dong, X. (2004). Systemic acquired resistance. Annual Review of Phytopathology, 42, 185–209.
Eckardt, N. A. (2008). Chitin signaling in plants: insights into the perception of fungal pathogens and rhizobacterial symbionts. Plant Cell, 20, 241–243.
El-Bebany, A. F., Rampitsch, C., & Daayf, F. (2010). Proteomic analysis of the phytopathogenic soilborne fungus Verticillium dahliae reveals differential protein expression in isolates that differ in aggressiveness. Proteomics, 10, 289–303.
Elvira, M. I., Galdeano, M. M., Gilardi, P., García-Luque, I., & Serra, M. T. (2008). Proteomic analysis of pathogenesis-related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum chinense L3 plants. Journal of Experimental Botany, 59, 1253–1265.
Fang, X., Jost, R., Finnegan, P. M., & Barbetti, M. J. (2013). Comparative proteome analysis of the strawberry-Fusarium oxysporum f. sp. fragariae pathosystem reveals early activation of defense responses as a crucial determinant of host resistance. Journal of Proteome Research, 12, 1772–1788.
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., & Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proceedings of the National Academy of Sciences of the United States of America, 100, 15253–15258.
Garcı́a-Limones, C., Hervás, A., Navas-Cortés, J. A., Jiménez-Dı́az, R. M., & Tena, M. (2002). Induction of an antioxidant enzyme system and other oxidative stress markers associated with compatible and incompatible interactions between chickpea (Cicer arietinum L.) and Fusarium oxysporum f. sp.ciceris. Physiological and Molecular Plant Pathology, 61, 325–337.
Ge, X., Chen, J., Li, N., Lin, Y., Sun, C., & Cao, K. (2003). Resistance function of rice lipid transfer protein LTP110. Biochem Mol Biol, 36, 603–607.
Glazebrook, J., Rogers, E. E., & Ausubel, F. M. (1997). Use of Arabidopsis for genetic dissection of plant defense responses. Annual Review of Genetics, 31(1), 547–569.
Grant, M. R., & Jones, J. D. G. (2009). Hormone (dis)harmony moulds plant health and disease. Science, 324, 750–752.
Hammond-Kosack, K. E., & Jones, J. D. (1996). Resistance gene-dependent plant defense responses. Plant Cell, 8, 1773–1791.
Hogenhout, S. A., Van der Hoorn, R. A. L., Terauchi, R., & Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Molecular Plant-Microbe Interactions, 22, 115–122.
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., & Nakai, K. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Research, 35(suppl 2): W585-W587.
Hou, M., Xu, W., Bai, H., Liu, Y., Li, L., Liu, L., Liu, B., & Liu, G. (2012). Characteristic expression of rice pathogenesis-related proteins in rice leaves during interactions with Xanthomonas oryzae pv. oryzae. Plant Cell Reports, 31, 895–904.
Hua, C., Wang, Y., Zheng, X., Dou, D., Zhang, Z., Govers, F., & Wang, Y. (2008). A Phytophthora sojae G-protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryotic Cell, 7, 2133–2140.
Jones, J. D. G., & Dangl, J. L. (2006). The plant immune system. Nature, 444, 323–329.
Kim, S. T., Cho, K. S., Jang, Y. S., & Kang, K. Y. (2001). Two-dimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis, 22, 2103–2109.
Koeck, M., Hardham, A. R., & Dodds, P. N. (2011). The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cellular Microbiology, 13(12), 1849–1857.
Legay, G., Marouf, E., Berger, D., Neuhaus, J. M., Mauch-Mani, B., & Slaughter, A. (2011). Identification of genes expressed during the compatible interaction of grapevine with Plasmopara viticola through suppression subtractive hybridization (SSH). European Journal of Plant Pathology, 129(2), 281–301.
Li, A., Meng, C., Zhou, R., Ma, Z., & Jia, J. (2006). Assessment of lipid transfer protein (LTP1) Gene in wheat powdery mildew resistance. Agricultural Sciences in China, 5, 241–249.
Lu, S., Friesen, T. L., & Faris, J. D. (2011). Molecular characterization and genomic mapping of the pathogenesis-related protein 1 (PR-1) gene family in hexaploid wheat (Triticum aestivum L.). Molecular Genetics and Genomics, 285, 485–503.
Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J., & Cameron, R. K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature, 419, 399–403.
Manickavelu, A., Kawaura, K., Oishi, K., Shin-I, T., Kohara, Y., Yahiaoui, N., Keller, B., Suzuki, A., Yano, K., & Ogihara, Y. (2010). Comparative gene expression analysis of susceptible and resistant near-isogenic lines in common wheat infected by Puccinia triticina. DNA Research, 17, 211–222.
Maytalman, D., Mert, Z., Baykal, A. T., Inan, C., Günel, A., & Semra, H. (2013). Proteomic analysis of early responsive resistance proteins of wheat (Triticum aestivum) to yellow rust (Puccinia striiformis f. sp. tritici) using ProteomeLab PF2D. Plant Omi, 6, 24–35.
McNeal, F. H., Konzak, C. F., Smith, E. P., Tate, W. S., & Russel, T. S. (1971). A uniform system for recording and processing cereal research data. Agric Res Serv Bull, 34-121.
Mellersh, D. G., Foulds, I. V., Higgins, V. J., & Heath, M. C. (2002). H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. The Plant Journal, 29, 257–268.
Mendoza, M. (2011). Oxidative burst in plant-pathogen interaction. Biotecnol Veg, 11, 67–75.
Nanda, A. K., Andrio, E., Marino, D., Pauly, N., & Dunand, C. (2010). Reactive oxygen species during plant-microorganism early interactions. Journal of Integrative Plant Biology, 52, 195–204.
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., & Jones, J. D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439.
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S., & Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nature Chemical Biology, 5, 308–316.
Rampitsch, C., Bykova, N. V., McCallum, B., Beimcik, E., & Ens, W. (2006). Analysis of the wheat and Puccinia triticina (leaf rust) proteomes during a susceptible host-pathogen interaction. Proteomics, 6, 1897–1907.
Rampitsch, C., Günel, A., Beimcik, E., & Mauthe, W. (2015). Proteome of monoclonal antibody-purified haustoria from Puccinia triticina race-1. Proteomics, 15(7), 1307–1315.
Rizhsky, L., Hallak-Herr, E., Van Breusegem, F., Rachmilevitch, S., Barr, J. E., Rodermel, S., Inze, D., & Mittler, R. (2002). Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. The Plant Journal, 32, 329–342.
Rovenich, H., Boshoven, J. C., & Thomma, B. P. (2014). Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Current Opinion in Plant Biology, 20, 96–103.
Scholtz, J. J. (2013). Identification of a putative protease inhibitor involved in three different puccinia-triticum aestivum interactions (doctoral dissertation, University Of The Free State Bloemfontein South Africa).
Selitrennikoff, C. P. (2001). Antifungal proteins. Applied and Environmental Microbiology, 67(7), 2883–2894.
Shiu, S.H., Bleecker, A.B., (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE, 113:re22.
Takakura, Y., CHE, F. S., Ishida, Y., Tsutsumi, F., KUROTANI, K. I., Usami, S., et al. (2008). Expression of a bacterial flagellin gene triggers plant immune responses and confers disease resistance in transgenic rice plants. Molecular Plant Pathology, 9(4), 525–529.
Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H. S., Han, B., et al. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen pseudomonas syringae. Plant Cell, 15(2), 317–330.
Tenberge, K. B., Beckedorf, M., Hoppe, B., Schouten, A., Solf, M., & von den Driesch, M. (2002). In situ localization of AOS in host-pathogen interactions. Microscopy and Microanalysis, 8, 250–251.
Tomich, M., Herfst, C. A., Golden, J. W., & Mohr, C. D. (2002). Role of flagella in host cell invasion by Burkholderia cepacia. Infection and Immunity, 70, 1799–1806.
Vaghefi, N., Mustafa, B. M., Dulal, N., Selby-Pham, J., Taylor, P. W. J., & Ford, R. (2013). A novel pathogenesis-related protein (LcPR4a) from lentil, and its involvement in defence against Ascochyta lentis. Phytopathologia Mediterranea, 52, 192–201.
van Loon, L. C., & Van Strien, E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiological and Molecular Plant Pathology, 55, 85–97.
van Loon, L. C., Rep, M., & Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology, 44, 135–162.
Vidhyasekaran, P. (2008). Perception and transduction of plant signals in pathogens. In P. Vidhyasekaran (Ed.), Fungal Pathogenesis in Plants and Crops: Molecular Biology and Host Defense Mechanisms (Second ed., ). Florida: CRC Press.
Walker, J. C. (1994). Structure and function of the receptor-like protein kinases of higher plants. Plant Molecular Biology, 26, 1599–1609.
Wang, X., Liu, W., Chen, X., Tang, C., Dong, Y., Ma, J., Huang, X., Wei, G., Han, Q., Huang, L., & Kang, Z. (2010). Differential gene expression in incompatible interaction between wheat and stripe rust fungus revealed by cDNA-AFLP and comparison to compatible interaction. BMC Plant Biology, 10, 9.
Wilm, M., Shevchenko, A., Houthaeve, T., Breit, S., Schweigerer, L., Fotsis, T., & Mann, M. (1996). Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature, 379, 466–469.
Xin, M., Wang, X., Peng, H., Yao, Y., Xie, C., Han, Y., Ni, Z., & Sun, Q. (2012). Transcriptome comparison of susceptible and resistant wheat in response to powdery mildew infection. Genomics, Proteomics & Bioinformatics, 10, 94–106.
Yang, A. H., & Yeh, K. W. (2005). Molecular cloning, recombinant gene expression, and antifungal activity of cystatin from taro (Colocasia esculenta cv. kaosiung no. 1). Planta, 221, 493–501.
Yao, R., Burr, D. H., Doig, P., Trust, T. J., Niu, H., & Guerry, P. (1994). Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Molecular Microbiology, 14, 883–893.
Zhou, H., Li, S., Deng, Z., Wang, X., Chen, T., Zhang, J., Chen, S., Ling, H., Zhang, A., Wang, D., & Zhang, X. (2007). Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. The Plant Journal, 52, 420–434.
Acknowledgments
This study was supported by TUBITAK, COST programme 109 T293 project. We thank Kadir Akan, Ayşe Yıldız and Lütfi Çetin for their help during plant inoculation and sampling. We also thank Abdulmecit Gökçe and Yavuz Öztürk for their technical support for PF2D as well as Konca Bulut and Rahmi Büyükkeskin for their experimental assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Demirci, Y.E., Inan, C., Günel, A. et al. Proteome profiling of the compatible interaction between wheat and stripe rust. Eur J Plant Pathol 145, 941–962 (2016). https://doi.org/10.1007/s10658-016-0882-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-016-0882-1