Abstract
Citrus canker, caused by the bacterial pathogen Xanthomonas citri subsp. citri (Xcc), is a serious leaf and fruit spotting disease affecting many important citrus cultivars including grapefruit and certain sweet oranges. Currently, efficacious and economical disease control measures for highly susceptible citrus cultivars are lacking. Development of commercial cultivars with greater field resistance to citrus canker is the optimum strategy for effective disease management. In this study, we generated transgenic ‘Duncan’ grapefruit (DG) and ‘Hamlin’ sweet orange (Ham) expressing the Arabidopsis NPR1 gene (AtNPR1), which is a key positive regulator of the long-lasting broad-spectrum resistance known as systemic acquired resistance (SAR). Our results indicate that over-expression of AtNPR1 in citrus increases resistance to citrus canker and that the resistance is related with the expression levels of AtNPR1 in the transgenic plants. The line (DG 42-2) with the highest expression level of AtNPR1 was also the most resistant, which developed significant fewer lesions accompanied by a ten-fold reduction in Xcc population. The lesions developed on DG 42-2 were smaller and darker than those on the control and lacked callus formation. These lesion phenotypes resemble those on canker resistant kumquats and canker susceptible citrus trees treated with SAR-inducing compounds. Therefore, over-expression of AtNPR1 in citrus is a promising approach for development of more resistant cultivars to citrus canker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asiatic citrus canker is a serious citrus disease caused by the bacterial pathogen, Xanthomonas citri subsp. citri (Xcc) that causes distinctive necrotic, erumpent lesions on leaves, stems, and fruits. Severe infections produce a range of symptoms that result in crop loss including defoliation, blemished fruit, and premature abscission and fruit drop (Graham et al. 2004). Many important commercial citrus cultivars including grapefruit (Citrus paradisi Macf.), certain sweet oranges (C. sinensis (L.) Osbeck), Key lime (C. aurantifolia Swingle), and lemons (C. limon (L.) Burm. F.), are moderately to highly susceptible to citrus canker (Gottwald et al. 1993). Reduced quality and quantity of fresh and processed fruit cause economic losses to the growers (Muraro et al. 2002). However, the most serious consequence is the impact on commerce resulting from restrictions to interstate and international fruit trade from canker-affected areas (Gottwald et al. 2009).
At present, management of citrus canker relies on both cultural practices and chemical control. Cultural practices include windbreaks and pruning or defoliation of diseased shoots to suppress the spread and severity of the disease (Behlau et al. 2008; Graham et al. 2004). Chemical control with sprays of copper-based bactericides reduces inoculum build-up on new leaf flushes and protects expanding fruit surfaces from infection, reducing incidence of blemished fruit and yield losses due to premature fruit drop (Stall et al. 1982; Graham and Leite 2004; Behlau et al. 2008). The efficacy of chemical control with copper-based bactericides depends on the susceptibility of the citrus cultivar, environmental conditions, and integration with other control measures (Leite and Mohan 1990). Although practices including windbreaks and copper bactericides reduce disease losses, these measures are costly (Muraro et al. 2002) and long-term copper use may lead to pathogen resistance (Rinaldi and Leite 2000) and significant accumulation in the soil environment (Alva et al. 1995).
Upon artificial inoculation of citrus leaf tissues, lesions caused by Xcc on susceptible cultivars develop hypertrophic and hyperplasic proliferation of cells resulting in raised, callus-like lesions on the surface. The highly resistant citrus relative, kumquat (Fortunella spp), develops very small brown to black lesions without callus or water-soaking. In kumquat, this reaction produces rapid necrosis and decline in populations of bacteria in reactions of up to 4 log units and therefore this has been described as hypersensitive response (HR)-like resistance (Francis et al. 2009a). Field susceptibility to Xcc varies widely among citrus types based on host range studies involving inoculation directly into the leaf mesophyll of immature tissues (Gottwald et al. 1993; Graham et al. 1990). To measure mesophyll tissue susceptibility of citrus cultivars to Xcc, inoculation of leaves and fruit without wounding is assessed by injection-infiltration of bacteria through the stomata (Viloria et al. 2004; Francis and Graham 2008). Based on in vitro and greenhouse assays to evaluate the host range and virulence of Xcc, reduction of lesions number, lesion size and bacterial populations by 10 fold or greater has been associated with field resistance to canker (Graham et al. 1990; Viloria et al. 2004).
The best long-term solution for management of citrus canker is to develop more resistance in commercially important citrus cultivars. Conventional approaches of citrus breeding for disease resistance in scion germplasm are difficult because resistance mainly occurs in citrus relatives such as kumquat whose genetic make-up interferes with the expression of optimum traits related to fruit quality and production (Viloria et al. 2004). Furthermore, conventional breeding is laborious, time-consuming and very expensive because citrus species are highly heterozygous, polygenic plants with a long juvenile period. In contrast, transgenic approaches can quickly incorporate resistance into citrus cultivars without interfering with the expression of optimum varietal traits. However, advance in citrus breeding through transgenic approaches is slowed by lack of molecular understanding of innate disease resistance in citrus.
In the model plant Arabidopsis thaliana, a natural disease resistance known as systemic acquired resistance (SAR) has been well characterized (Durrant and Dong 2004). SAR is an inducible defence mechanism that confers plants long-lasting resistance against a broad spectrum of pathogens. The defence signal molecule salicylic acid (SA) is an essential and sufficient signal molecule for the activation of SAR. Exogenous application of SA or its biologically active analogs, such as 2,6-dichloroisonicotinic acid (INA) and benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH), leads to the activation of SAR; whereas prevention of SA accumulation by genetic mutations compromises the induction of SAR (Wildermuth et al. 2001). In citrus, foliar or soil application of SA analogs, such as INA, acibenzolar-S-methyl (ASM, Actigard®, Syngenta Crop Protection), and imidacloprid (Imid, Admire®, Bayer Crop Science), induces the expression of the pathogenesis-related (PR) gene, PR-2 (used as a marker gene of SAR in citrus, encoding β-1,3 glucanase) (Dekkers et al. 2004; Francis et al. 2009b), and produces protection against citrus canker under greenhouse conditions and in field trials (Francis et al. 2009b; Graham and Myers 2009). These results demonstrate that the SA-mediated SAR is effective for control of citrus canker in the field.
The Arabidopsis NPR1 gene (AtNPR1) has been well established as a key positive regulator of SAR, which acts downstream of the signal molecule SA. Mutations in NPR1 not only block SA-induced PR gene expression and SAR, but also increase susceptibility to pathogens (Cao et al. 1994). In contrast, over-expression of AtNPR1 in transgenic Arabidopsis enhances resistance to bacterial and oomycete pathogens (Cao et al. 1998). The enhanced resistance is associated with a faster or stronger response of the transgenic plants to pathogen infection and the SAR signal molecule SA (Friedrich et al. 2001). Over-expression of the AtNPR1 gene or its orthologs also enhances disease resistance in many crop plants including rice, wheat, rapeseed, tomato, and apple (Lin et al. 2004; Chern et al. 2005; Makandar et al. 2006; Malnoy et al. 2007; Potlakayala et al. 2007). Interestingly, over-expression of AtNPR1 in most plant species does not cause obvious detrimental effects on plant growth and development (Cao et al. 1998; Lin et al. 2004), which makes AtNPR1 a workable target for genetic engineering of nonspecific resistance in plants.
In this work, we took a transgenic approach to generating ‘Duncan’ grapefruit and ‘Hamlin’ sweet orange plants expressing AtNPR1. Characterization of the transgenic plants indicates that expression of AtNPR1 in citrus is a highly effective means to control citrus canker.
Materials and methods
Plasmid construction, plant transformation, and SA measurement
The full-length cDNA of AtNPR1 was removed from pKEx4tr-AtNPR1 by digestion with EcoR I plus Sac I and cloned into EcoR I/Sac I-digested pBI1.4T vector. The resulting plasmid pBI1.4T-AtNPR1 was introduced into Agrobacterium strain EHA105 by electroporation, and transformed into ‘Duncan’ grapefruit and ‘Hamlin’ sweet orange following the published protocol by the transformation laboratory at Citrus Research and Education Center (CREC) of the University of Florida (Orbović and Grosser 2006). Measurement of SA in plant tissues was done by HPLC as described by Verberne et al. (2002).
Pathogen infection
Bacterial inoculum was prepared as previously described (Francis et al. 2009b). Briefly, Xcc strain X2002-0014 was cultured in nutrient broth and grown at 28°C for 24 h to log phase. Bacterial suspension was centrifuged at 10,000 g for 20 min, re-suspended in PBS, and adjusted to 0.1 OD at A620nm equivalent to 108 colony-forming units (cfu) ml-1. Bacterial cell density was adjusted to 105 cfu ml-1 for inoculation.
Evaluation of citrus leaf resistance was conducted in vitro as previously described (Francis et al. 2009b). For this assay, immature leaves (75% expanded) were collected in the morning from greenhouse seedlings of the transgenic plants and placed inside sealable plastic bags on ice for transport to the lab. Leaf surfaces were disinfected by dipping the leaves in 0.5% sodium hypochlorite for 30 s and immediately rinsed 3 times with sterile water. Using a 1-ml needleless syringe, the bacterial suspension was pressure infiltrated into the abaxial leaf surface until the water-soaked area reached about 6 mm in diameter. Three injection infiltrations were performed on each side of the mid-vein. The inoculated leaves were placed on soft water agar (0.5%) surface in 15 cm diameter Petri dish plates with the abaxial side up, and pressed onto the agar surface with a plastic spreader to obtain as much contact as possible with the agar surface. Petri dishes were immediately sealed with parafilm and the plates were incubated in an environmentally controlled growth chamber at 28°C under fluorescent light (60 μEinsteins s-1 m-2 for 12-hr photoperiods). Lesions were counted at each inoculation site 14 days after inoculation. Bacterial populations in the inoculated areas were estimated 14 days post-inoculation. Leaf discs (6 mm diameter) were excised from the infiltrated area and ground with 1.0 ml of PBS buffer using a glass homogenizer. Serial dilutions of suspension were plated on kasugamycin-cephalexin-chlorothalonil (KCC; nutrient agar plus kasugamycin 16.0 mgl-1, cephalexin 16.0 mg l-1, and chlorothalonil 12.0 mgl-1) agar medium (Viloria et al. 2004). Total bacterial colonies per inoculation site were expressed as log cfu ml-1. The numbers of lesions and bacterial populations were analyzed statistically (SAS Institute Inc., Cary, NC, 1996) using ANOVA in nested design. Significant differences were calculated with Waller ’s K-ratio t-test for pair-wise comparisons.
PCR and real-time quantitative PCR analysis
Citrus genomic DNA was extracted following the previously described method (Cheng et al. 2003). The AtNPR1 transgene was amplified by PCR using the primers NPR1F (5’-ATGGACACCACCATTGATGG-3’) and NPR1R (5’-TCACCGACGACGATGAGAGAG-3’). The PCR reactions were performed under the following conditions: 94°C for 3 min, 35 cycles (94°C for 1 min, 56°C for 1 min, 72°C for 2 min), and a final extension at 72°C for 10 min. Citrus RNA was extracted using an RNeasy® Plant Mini Kit (QIAGEN Sciences Inc., Germantown, MD) following the manufacturer’s instructions. Reverse transcription (RT) and real-time quantitative PCR (Q-PCR) were performed as previously described (Francis et al. 2009b). Reactions were run and analyzed on a MX3000P real-time PCR machine (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. The relative quantity of AtNPR1 and PR2 is expressed in relation to elongation factor-1-alpha (EF1α) using the formula 2^(Ct[EF1α]-Ct[gene]), where 2 represents perfect PCR efficiency. The primers used for AtNPR1 were Q-NPR1F (5’-AGCATTCTCTCAAAGGCCGAC-3’) and Q-NPR1R (5’-TGAGACGGTCAGGCTCGAGG-3’), and those for PR2 and EF1α were described previously (Mozoruk et al. 2006; Francis et al. 2009b).
Results
Generation of citrus transgenic plants expressing the Arabidopsis NPR1 gene
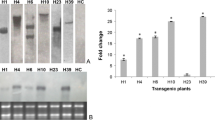
‘Duncan’ grapefruit and the sweet orange ‘Hamlin’ were chosen as recipients for genetic transformation because they are among the most susceptible commercial cultivars to citrus canker (Stall et al. 1980). The full-length cDNA of AtNPR1 was cloned in between the cauliflower mosaic virus (CaMV) 35 S promoter and the nopaline synthase (NOS) terminator of the plant binary vector pBI1.4T. The resulting plasmid pBI1.4T-AtNPR1 was introduced into Agrobacterium tumefaciens. ‘Duncan’ grapefruit and ‘Hamlin’ hypocotyls were transformed using Agrobacteria harbouring pBI1.4T-AtNPR1. Nine independent ‘Duncan’ grapefruit (DG) transgenic lines and seven independent ‘Hamlin’ (Ham) transgenic lines were generated. The presence of AtNPR1 in the nine DG transgenic lines and six Ham transgenic lines was confirmed by PCR analysis using AtNPR1 specific primers (Fig. 1a). Although the AtNPR1 gene was not detected in the transgenic line Ham 13–60, this line was included in all experiments as a control.
Generation of citrus transgenic plants expressing the AtNPR1 gene. a PCR analysis of the presence of the AtNPR1 gene in the transgenic plants. The AtNPR1 gene (1,782 bp, top panel) was detected in the nine ‘Duncan’ grapefruit (DG) transgenic lines and six ‘Hamlin’ (Ham) transgenic lines, but not in the non-transformed controls (DG/Ham No Trans) and one transgenic line (Ham 13–60). The citrus EF1α gene (bottom panel) was used as a control showing the presence of citrus genomic DNA in the PCR reactions. b Q-PCR analysis of the expression levels of the AtNPR1 gene in different transgenic lines. The transgenic lines were arbitrarily placed into three groups: high (H), middle (M), and low (L), based on the expression levels of the AtNPR1 gene
We then tested the expression levels of AtNPR1 in the transgenic plants. Expression levels of the AtNPR1 transgene varied widely among the 16 independent transgenic lines (Fig. 1b). Based on the relative expression levels of AtNPR1, the transgenic lines were classified into three groups: high (H), middle (M), and low (L) (Fig. 1b). Line DG 42-2, sole member of the H group, expressed a dramatically higher level of AtNPR1 than the M lines, whereas the L lines expressed significantly lower levels of AtNPR1 than the M lines. Background levels of AtNPR1 transcripts were not detected in the non-transformed controls (DG/Ham No Trans), confirming the specificity of the primers used for the Q-PCR analysis.
Resistance of the transgenic plants to citrus canker
To test whether over-expression of AtNPR1 confers resistance to citrus canker, leaves of the transgenic plants were inoculated in vitro with Xcc. Symptoms were characterized on day 14 and day 30 post-inoculation (Fig. 2). On day 14, the non-transformed ‘Duncan’ grapefruit (DG No Trans) and DG L lines produced callus-like lesions, whereas the H line DG 42-2 developed fewer necrotic lesions that were smaller in size. The DG M lines such as DG 57-28 developed an intermediate lesion phenotype. The non-transformed ‘Hamlin’ (Ham No Trans) and Ham L lines produced numerous callused lesions, whereas the Ham M lines such as Ham 13-3 exhibited an HR-like reaction with cell collapse in the inoculated area at 24 h post-inoculation that persisted without development of callus tissue. On day 30, the non-transformed ‘Duncan’ grapefruit and DG L lines produced callus-like lesions that were suberized and surrounded by a chlorotic halo, whereas the H line DG 42-2 produced necrotic lesions that did not develop into callus-like cankers. The non-transformed ‘Hamlin’ and Ham L lines produced callused lesions that were suberized with water-soaked margins surrounded by a chlorotic halo. The Ham M lines still showed tissue collapse with little callus proliferation.
Phenotype of the citrus canker lesions on the transgenic plants. Leaves were inoculated by injection infiltration with a suspension of Xcc and lesion development was assessed 14 and 30 days after inoculation. The non-transformed controls (DG/Ham No Trans) developed typical canker lesions, whereas the transgenic plants developed fewer lesions with altered morphology. dpi: day post-inoculation
The resistance produced by the AtNPR1 transgene was also reflected by the numbers of lesions developed on the inoculated leaves (Table 1). For both ‘Duncan’ grapefruit and ‘Hamlin’, the lesion numbers on the M lines (except DG 48-2) were significantly lower than those on the controls, whereas the lesion numbers on the L lines were not significantly different from those on the controls. The most resistant H line DG 42-2 had an average of 10.8 lesions per leaf disc compared to 59.4 lesions per leaf disc for the non-transformed control.
Xcc populations in lesions on day 14 post-inoculation were also reduced in the transgenic ‘Duncan’ grapefruit expressing AtNPR1 (Table 1). Both DG M lines and the H line had a significantly lower Xcc population than the control. The most resistant H line DG 42-2 produced a more than one-log-unit reduction in Xcc population compared with the control. In contrast, compared with the ‘Hamlin’ control, Ham M lines that had a reduced number of lesions did not show a reduction in Xcc population.
SA levels in the transgenic plants
Over-expression of AtNPR1 in plants like rice that contain high basal levels of SA leads to detrimental effects on plant growth and development (Silverman et al. 1995; Fitzgerald et al. 2004; Chern et al. 2005). Although growth reductions or abnormalities were not observed for the transgenic lines of ‘Duncan’ grapefruit and ‘Hamlin’, it is not known whether citrus plants accumulate high basal levels of SA, and whether over-expression of AtNPR1 changes the SA levels in citrus plants. To this end, levels of free SA and total SA (SA + SAG) were assayed in non-transformed ‘Duncan’ grapefruit and ‘Hamlin’ as well as the transgenic lines. As shown in Fig. 3, the SA levels in both ‘Duncan’ grapefruit and ‘Hamlin’ were very low and comparable with those in Arabidopsis plants without pathogen infection. Furthermore, over-expression of AtNPR1 did not alter the basal levels of SA in the transgenic plants.
Salicylic acid levels in the transgenic plants. HPLC analysis of free SA a and total SA (SA + SAG) b in the transgenic plants. The non-transformed controls (DG/Ham No Trans) did not accumulate high basal levels of SA and over-expression of AtNPR1 did not change the SA levels in the transgenic plants. Values represent the average of three biological replicates with standard deviation. The experiment was repeated with similar results. Arabidopsis plants with or without pathogen infection were used as controls. FW: fresh weight. At: Arabidopsis thaliana. Psm: Pseudomonas syringae pv. maculicola ES4326
PR2 gene expression in the transgenic plants
Over-expression of AtNPR1 in rice leads to a constitutive defence response and causes spontaneous lesion development (Fitzgerald et al. 2004; Chern et al. 2005). Although the morphology of the transgenic citrus plants expressing AtNPR1 appears to be normal, it was not clear whether the transgenic plants exhibit a constitutive defence response. To test this, we monitored the expression levels of the PR2 gene (the SAR marker gene in citrus) in the transgenic plants. The expression levels of PR2 in all transgenic lines were comparable with those in the non-transformed ‘Duncan’ grapefruit and ‘Hamlin’ plants (Fig. 4), suggesting that over-expression of AtNPR1 in citrus may not induce a constitutive defence response.
Expression levels of the PR2 gene in the transgenic plants. Q-PCR analysis of the expression levels of PR2 in the transgenic plants. Over-expression of AtNPR1 did not change the expression levels of PR2 in the transgenic plants. Values represent the average of three biological replicates with standard deviation. The experiment was repeated with similar results
Discussion
The AtNPR1 gene is a key positive regulator of the long-lasting SAR, which has been utilized in many crop plants to improve their resistance to pathogens (Lin et al. 2004; Chern et al. 2005; Makandar et al. 2006; Malnoy et al. 2007; Potlakayala et al. 2007). Consistent with the reports in other crop plants, over-expression of AtNPR1 in citrus conferred heightened resistance to citrus canker and the level of resistance was correlated with the amounts of the AtNPR1 transcripts. In the ‘Duncan’ grapefruit transgenic line DG 42-2 that expressed the highest levels of AtNPR1, the reduction of the callus phenotype and the size of the lesions were similar to those obtained for HR-resistant kumquat inoculated by similar methods in vitro as used in the current study (Francis, M. I., Peña, A., & Graham, J. H., submitted to EJPP). In this line, the phenotype produced in vitro is associated with a one-log-unit reduction in Xcc populations in the lesions, which represents a high level of resistance when considered in the context of resistance of citrus cultivars in the field (Graham et al. 1990). These results indicate that AtNPR1 is highly effective for activation of resistance and reduction of the bacterial multiplication and lesion severity in vitro. Interestingly, compared with the non-transformed controls, the ‘Duncan’ grapefruit AtNPR1-expressing lines with a reduced number of lesions generally showed a reduction in Xcc population, whereas the ‘Hamlin’ AtNPR1-expressing lines did not show any reduction in Xcc population. This discrepancy between ‘Duncan’ grapefruit and ‘Hamlin’ may be due to the differences in levels of resistance expression in the two genotypes.
Over-expression of AtNPR1 in rice leads to spontaneous development of chlorotic lesions, cell death, accumulation of hydrogen peroxide, and constitutive expression of defence genes (Chern et al. 2005). BTH treatment enhances these phenotypes, indicating that the detrimental phenotypes are likely a result of a constitutive defence response in the transgenic plants (Fitzgerald et al. 2004). Because rice contains high basal levels of SA (an order of magnitude higher than that found in other plant species) (Silverman et al. 1995), these phenotypes may be caused by the constitutive expression of NPR1-dependent defence pathways in the rice plants over-expressing AtNPR1. We found that both non-transformed ‘Duncan’ grapefruit and ‘Hamlin’ plants had low basal levels of SA. Expression of AtNPR1 in ‘Duncan’ grapefruit and ‘Hamlin’ did not alter the SA levels. Furthermore, the transgenic plants did not constitutively express the citrus SAR marker gene, PR2. These results together suggest that over-expression of AtNPR1 in citrus may not induce constitutive defence responses. The heightened resistance is likely due to a faster or stronger response of the transgenic plants to pathogen infection (Friedrich et al. 2001). Therefore, over-expression of AtNPR1 in citrus may not interfere with normal plant growth and development. Consistent with this is that no abnormal phenotypes for leaves and shoots of the transgenic lines were observed. However, further field investigations will be required to evaluate the effect of over-expression of AtNPR1 on horticultural traits related to tree vigour, fruit quality and production of the transgenic plants.
In Arabidopsis, over-expression of AtNPR1 leads to increased fungicide efficacy (Friedrich et al. 2001). Suboptimal rates of fungicides that marginally control infection of wild-type plants substantially reduce fungal disease in AtNPR1-overexpressing plants. Since the AtNPR1-overexpressing citrus plants in this study are not immune to citrus canker, disease management with transgenic resistance will still need to be integrated with chemical control. Increased bactericide efficacy should reduce the rate and frequency of applications of copper-based bactericides for disease control. Reduction in amount of copper for effective canker control will reduce soil accumulation of copper and may decrease the selection pressure for development of copper resistance (Rinaldi and Leite 2000). Additionally, over-expression of AtNPR1 in other crops can protect plants from diverse pathogens including bacterial and fungal pathogens (Lin et al. 2004; Chern et al. 2005; Makandar et al. 2006; Malnoy et al. 2007; Potlakayala et al. 2007). Whether the AtNPR1-overexpressing citrus plants are more resistant to other pathogens awaits further investigation.
Although it has been shown that SA analogs induce disease resistance in citrus (Dekkers et al. 2004; Francis et al. 2009b; Graham and Myers 2009), the role of SA and SAR in citrus immunity has not been established. The fact that over-expression of AtNPR1, a key regulator of SAR, increases disease resistance in citrus suggests that the SAR signalling pathway exists in this plant. The identity of NPR1 ortholog(s) in citrus has yet to be revealed and whether over-expression of citrus NPR1 in citrus can increase disease resistance needs to be determined. Identification and characterization of citrus NPR1 will facilitate generation of disease resistant citrus plants using a cisgenic approach (Schouten et al. 2006), which may help avoid genetic modification issues in the near future.
Abbreviations
- SA:
-
salicylic acid
- AtNPR1:
-
Arabidopsis nonexpressor of pathogenesis-related genes 1
- SAR:
-
systemic acquired resistance
- PR-2 :
-
pathogenesis-related protein 2
- INA:
-
2,6-dichloroisonicotinic acid
- BTH:
-
benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester
- ASM:
-
acibenzolar-S-methyl
- Imid:
-
imidacloprid
- Xcc :
-
Xanthomonas citri ssp. citri
- Psm :
-
Pseudomonas syringae pv. maculicola
- HR:
-
hypersensitive response
References
Alva, A. K., Graham, J. H., & Anderson, C. A. (1995). Soil pH and copper effects on young '‘Hamlin’' orange trees. Soil Science Society of America Journal, 59, 481–487.
Behlau, F., Belasque, J., Jr., Bergamin Filho, A., Graham, J. H., Leite, R. P., Jr., & Gottwald, T. R. (2008). Copper sprays and windbreaks for control of citrus canker on young orange trees in southern Brazil. Crop Protection, 27, 807–813.
Cao, H., Bowling, S. A., Gordon, S., & Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell, 6, 1583–1592.
Cao, H., Li, X., & Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proceedings of the National Academy of Sciences of the United States of America, 95, 6531–6536.
Cheng, Y., Guo, W., Yi, H., Pang, X., & Deng, X. (2003). An efficient protocol for genomic DNA extraction from citrus species. Plant Molecular Biology Reporter, 21, 177a–177g.
Chern, M., Fitzgerald, H. A., Canlas, P. E., Navarre, D. A., & Ronald, P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Molecular Plant-Microbe Interactions, 18, 511–520.
Dekkers, M. G. H., Graham, J. H., Burns, J. K., Cubero, J., & Colburn, G. C. (2004). Evaluation of chemical inducers and PR protein reporters for induced systemic resistance to citrus bacterial diseases. Phytopathology, 94, S25.
Durrant, W. E., & Dong, X. (2004). Systemic acquired resistance. Annual Review of Phytopathology, 42, 185–209.
Fitzgerald, H. A., Chern, M.-S., Navarre, R., & Ronald, P. C. (2004). Over-expression of (At)NPR1 in rice leads to a BTH- and environment-inducible lesion-mimic/cell death phenotype. Molecular Plant-Microbe Interactions, 17, 140–151.
Francis, M. I., & Graham, J. H. (2008). In vitro inoculation of citrus germplasm for rapid screening of resistance to citrus canker. Phytopathology, 98, S55.
Francis, M. I., Peña, A., Kostenyuk, I., Burns, J., & Graham, J. H. (2009a). HR-like resistance of kumquat (Fortunella spp.) to citrus canker caused by Xanthomonas citri sbsp. citri. Phytopathology, 99, S36.
Francis, M. I., Redondo, A., Burns, J. K., & Graham, J. H. (2009b). Soil application of imidacloprid and related SAR-inducing compounds produces effective and persistent control of citrus canker. European Journal of Plant Pathology, 124, 283–292.
Friedrich, L., Lawton, K., Dietrich, R., Willits, M., Cade, R., & Ryals, J. (2001). NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Molecular Plant-Microbe Interactions, 14, 1114–1124.
Gottwald, T. R., Graham, J. H., Civerolo, E. L., Barrett, H. C., & Hearn, C. J. (1993). Differential host range reaction of citrus and citrus relatives to citrus canker and citrus bacterial spot determined by leaf mesophyll susceptibility. Plant Disease, 77, 1004–1009.
Gottwald, T., Graham, J., Bock, C., Bonn, G., Civerolo, E., Irey, M., et al. (2009). The epidemiological significance of post-packinghouse survival of Xanthomonas citri ssp. citri for dissemination of Asiatic citrus canker via infected fruit. Crop Protection, 28, 508–524.
Graham, J. H., & Leite, R. P., Jr. (2004). Lack of control of citrus canker by induced systemic resistance compounds. Plant Disease, 88, 745–750.
Graham, J. H., & Myers, M. (2009). Soil drenches of imidacloprid, thiamethoxam and acibenzolar-S-methyl for induction of SAR to control citrus canker in young citrus trees. Phytopathology, 99, S46.
Graham, J. H., Gottwald, T. R., & Fardelmann, D. (1990). Cultivar-specific interactions for strains of Xanthomonas campestris from Florida that cause citrus canker and citrus bacterial spot. Plant Disease, 74, 753–756.
Graham, J. H., Gottwald, T. R., Cubero, J., & Achor, D. S. (2004). Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Molecular Plant Pathology, 5, 1–15.
Leite, R. P., Jr., & Mohan, S. K. (1990). Integrated management of citrus canker disease caused by Xanthomonas campestris pv. citri in the State of Parana, Brazil. Crop Protection, 9, 3–7.
Lin, W. C., Lu, C. F., Wu, J. W., Cheng, M. L., Lin, Y. M., Yang, N. S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Research, 13, 567–581.
Makandar, R., Essig, J. S., Schapaugh, M. A., Trick, H. N., & Shah, J. (2006). Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Molecular Plant-Microbe Interactions, 19, 123–129.
Malnoy, M., Jin, Q., Borejsza-Wysocka, E. E., He, S. Y., & Aldwinckle, H. S. (2007). Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Molecular Plant-Microbe Interactions, 20, 1568–1580.
Mozoruk, J., Hunnicutt, L. E., Cave, R. D., Hunter, W. B., & Bausher, M. G. (2006). Profiling transcriptional changes in Citrus sinensis (L.) Osbeck challenged by herbivory from the xylem-feeding leafhopper Homalodisca coagulata (Say) by cDNA macroarray analysis. Plant Science, 170, 1068–1080.
Muraro, R. P., Roka, F. M., & Spreen, T. H. (2002). Grower costs of having citrus canker in Florida with an overview of Argentina’s citrus canker control program. Staff Paper SP02-3. Department of Food Resource Economics, Institute of Food and Agricultural Science, University of Florida
Orbović, V., & Grosser, J. W. (2006). Citrus: Sweet orange (Citrus sinesis L. Osbeck 'Valencia') and Carrizo citrange (Citrus sinesis (L.) Osbeck x Poncirus trifoliata (L.) Raf.). In K. Wang (Ed.), Agrobacterium protocol-methods in molecular biology (pp. 177–189). Totowa: Humana.
Potlakayala, S. D., DeLong, C., Sharpe, A., & Robert, P. R. (2007). Conservation of non-expressor of pathogenesis-related genes1 function between Arabidopsis thaliana and Brassica napus. Physiological and Molecular Plant Pathology, 71, 174–183.
Rinaldi, D. A. M. F., & Leite, R. P., Jr. (2000). Adaptation of Xanthomonas axonopodis pv. citri population to the presence of copper compounds in nature. Proceedings of the International Society of Citriculture, 2, 1064.
Schouten, H. J., Krens, F. A., & Jacobsen, E. (2006). Cisgenic plants are similar to traditionally bred plants. EMBO Reports, 7, 751–753.
Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Métraux, J.-P., & Raskin, I. (1995). Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiology, 108, 633–639.
Stall, R. E., Miller, J. W., Marco, G. M., & Canteros de Echenique, B. I. (1980). Population dynamics of Xanthomonas citri causing cancrosis of citrus in Argentina. Proceedings of the Florida State Horticultural Society, 93, 10–14.
Stall, R. E., Miller, J. W., Marco, G. M., & Canteros de Echenique, B. I. (1982). Timing of sprays to control cancrosis of grapefruit in Argentina. Proceedings of the International Society of Citriculture, 1, 141–417.
Verberne, M. C., Brouwer, N., Delbianco, F., Linthorst, H. J., Bol, J. F., & Verpoorte, R. (2002). Method for the extraction of the volatile compound salicylic acid from tobacco leaf material. Phytochemical Analysis, 13, 45–50.
Viloria, Z., Drouilard, D. L., Graham, J. H., & Grosser, J. W. (2004). Screening triploid hybrids of 'Lakeland' Limequat for resistance to citrus canker. Plant Disease, 88, 1056–1060.
Wildermuth, M. C., Dewdney, J., Wu, G., & Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565.
Acknowledgments
This work was supported by grants from Florida Citrus Production Research Advisory Council (FCPRAC) awarded to Z.M., W.D., and J.H.G. and a grant from the National Science Foundation (IOS-0842716) awarded to Z.M.. The authors are grateful to Dr. Xinnian Dong (Duke University) for the pKEx4tr-AtNPR1 cDNA plasmid and Dr. Sixue Chen (University of Florida) for access to the HPLC equipment. The authors thank Cecile Robertson and Alma Peña for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, X., Francis, M.I., Dawson, W.O. et al. Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur J Plant Pathol 128, 91–100 (2010). https://doi.org/10.1007/s10658-010-9633-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-010-9633-x