Abstract
Pollution is one of the main concerns in marine ecosystems nowadays. Environmental anthropogenic-mediated toxicants may affect genetic diversity both at the individual and ecosystem levels and may also alter the genetic structure of populations. This study examined the temporal pattern of genetic diversity among populations of the benthic bivalve Ensis siliqua in two locations of Galicia, following the Prestige oil spillage. On November 13, 2002 the oil tanker Prestige sank at 240 km from Galician coast and 63,000 tonnes of heavy fuel were released to the marine environment. E. siliqua samples were sampled between 2001 and 2006. Genetic variation was assessed by means of Random Amplification of Polymorphic DNA (RAPD). A significant decrease in genetic diversity was observed for the 2006 samples. Nei’s genetic distance, fixation index (PhiPT), and PCA values also supported differences in the 2006 samples. We hypothesize that the temporal genetic variation observed in E. siliqua populations is due to a strong effect of genetic drift caused by a reduction in population size and that the indirect effects of the Prestige spill possibly caused this reduction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Environmental toxicants, mainly those produced by anthropogenic activities, may induce genetic alterations in aquatic ecosystems. Pollution is known to affect the genetic structure of natural populations (Belfiore and Anderson 2001; Matson et al. 2006), either through a direct mutagenic effect of the toxicant or via contaminant-mediated mortality and/or alteration in physiologic or reproductive parameters (e.g., Bickham et al. 2000; Theodorakis et al. 2001; de Wolf et al. 2004a). This is particularly true for species with short generation times and/or with a sessile period of their life including fish, amphibians and many invertebrates (Matson et al. 2006; Chung et al. 2010). Environmental contaminants can lead to a reduction in genetic diversity resulting from strong selection for chemical tolerance, from an increased mortality, from founder effects resulting from re-establishment of extinct populations or as a result of a reduction in the number of breeding adults (Nadig et al. 1998; Matson et al. 2006).
To assess the impact of pollution in marine communities, indicators such as diversity index or population densities are often used (see Johnston and Roberts 2009). Nevertheless, these indicators tend to recover their initial values shortly after removal of pollution and do not reflect the possible alteration of population gene composition, making the evaluation of pollution effects difficult (Bickham et al. 2000). On the other hand, population genetic analyses offer powerful and alternative tools to evaluate pollutant-mediated effects, since these analyses examine the current genetic population diversity and may detect alterations in genetic diversity in exposed populations (Theodorakis et al. 2001; De Wolf et al. 2004a). Nonetheless, the possibility to foresee future directions of populations from genetic analyses must be viewed with caution particularly when genetic drift is a major determinant (De Wolf et al. 2004a). In this sense, the study of temporal genetic variation of population structure is essential to fully understand factors (including anthropogenic stressors) that affect genetic variability and demographic processes within species (Avise 2000; Palumbi 2004; Calderón et al. 2009).
Catastrophic events such as tanker wrecks release large amounts of pollutants into the marine environment. On November 13, 2002 the oil tanker Prestige sank at 240 km off the Galician coast (NW Spain) and spilled about 63,000 tonnes of heavy fuel oil (Figueras et al. 2005). Several thousands of kilometres of the Spanish Atlantic littoral were affected. The Galician region was the most disturbed, with most of it coast (approximately 70% according to the International Oil Pollution Compensation Funds) being affected by the oil spill. Nevertheless, the degree and intensity of the pollution showed important differences among areas of Galicia due to local hydrogeographic features and the characteristics of the fuel (low solubility, high viscosity, and slow degradation).
Marine organisms, especially those with low mobility or sessile phases might have suffered more acutely the toxic effects of the oil spill. This might be the case of the razor clam Ensis siliqua, a bivalve with a great economic value that appears along the Northwest Spain where it often forms extensive dense beds (Darriba et al. 2005). This species is most abundant at water depths of 3 and 7 m, although there are registers of specimens found at 10 m (Monteiro and Gaspar 1993) or even 50 m depth (von Cosel 1990). Ensis siliqua is a dioecious (gonochoric or unisexual) species with a reproductive cycle that consists of a long sexual rest period in summer and autumn, a gametogenesis period in November and December, and with an only spawning period occurring in April–May (Darriba et al. 2005). First maturation in E. siliqua is different depending on latitude and climate situation, with Southern Portuguese specimens maturing in its first year (Gaspar and Monteiro 1998) meanwhile maturity is reached at 3 years of age in North Wales and Northern Ireland populations (Fahy and Gaffney 2001). Regarding the age structure of razor clam beds, it is usual that unexploited ones consist of older animals (up to 25 years old) whereas exploited ones are a mixture of small (younger) and large (older) animals. Densities of E. siliqua individuals on beds are usually high, with some studies reporting up to 200 specimens/m2 (Fahy and Gaffney 2001). Razor clams are highly active and mobile; they burrow very fast and can use foot for propulsion, being thus able to select more favourable substratum and growing conditions. The planktonic phase of E. siliqua larvae is mobile, and shows ability to migrate in the water column, and with a residence time in water column of about 14–15 days (Da Costa et al. 2010). Thus, it is assumed that E. siliqua larvae have a potential for long-distance dispersal.
RAPD analysis is one of the molecular techniques used in assaying population genetic studies that has recently been used successfully to carry out ecotoxicological studies (Theodorakis et al. 2001; Theodorakis and Bickham 2004, Liu et al. 2007; Körpe and Aras 2011). The advantages of RAPD technique include (a) the analysis of a large number of segments among the genome, (b) the applicability to unstudied species, and (c) the easy performance. Furthermore, it has been conveniently applied to study the genetic structure of several Atlantic populations of E. siliqua (Fernández-Tajes et al. 2007; Arias et al. 2010).
The aim of this study was therefore to assess the temporal stability of the level of genetic diversity and population differentiation among samples of the economically important razor clam E. siliqua from two locations of the Galician coast affected by the Prestige oil spill. Samples were taken before and after the Prestige incident and were evaluated employing RAPD markers.
Materials and methods
Sample collection and DNA extraction
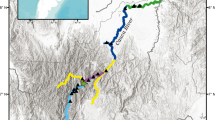
Specimens of E. siliqua were obtained by diving from two locations in the Galicia coast, Fisterra (Fis) and Barra de Cangas (Bar) (Fig. 1), on four dates 2001 (FisPr, BarPr), 2004 (Fis04, Bar04), 2005 (Fis05, Bar05), and 2006 (Fis06, Bar06). Immediately after collection, the razor clams were transported alive to the laboratory, dissected and preserved in 95% ethanol. DNA was isolated according to the protocol described by Fernández-Tajes et al. (2007).
RAPD amplification
The amplification and visualization of 61 RAPD loci obtained with five primers was performed using the methodology described by Fernández-Tajes et al. (2007): PCR reaction mixture was carried out in 25 μl of reaction volume containing 25 ng genomic DNA, 1.25 U Taq DNA polymerase, 0.2 mM dNTPs, 0.2 μM each tested primer and 1.75 mM MgCl2.). The PCR reaction was carried out in a MJC-Thermocycler (BioRad) with the following program: an initial denaturation cycle of 3 min at 94°C, followed by ten low astringency cycles at 94°C for 45 s, 40°C for 1 min, and 72°C for 75 s, then, 30 high astringency cycles at 94°C for 45 s, 50°C for 1 min, and 72°C for 1 min. A final extension was carried out at 72°C for 5 min. To ensure reproducibility, RAPD patterns were tested for the effect of Mg2+, DNA, dNTP and Taq DNA polymerase concentrations, which can affect the size and the number of amplified products.
RAPD band selection and scoring
Primer and band selection were performed in Pre-Prestige samples. The selection approaches of the bands-pattern were: the amplification quality, the non-coincidence with the negative control (reaction mixture without DNA) and the reproducibility. The RAPD markers were coded in function of the primer name and the size of each band. Only those bands with a frequency inferior to the following relationship. P = 1 − (3/N), P being the observed frequency and N the number of individuals per population, were selected for analyses (Lynch and Milligan 1994).
All the amplifications were made at least twice to check for reproducibility in the selected bands. During all the study, all reactions were carried out in the same thermocycler (MJC-Thermocycler, BioRad). A positive control (two DNA samples chosen from preliminary primer selection and successfully amplified in previous studies (Fernández-Tajes et al. 2007; Arias et al. 2010) and two negative controls (no DNA template) were incorporated in all reactions. Additionally, two samples corresponding to different sample years from current amplification batch were also incorporated in every amplification sets in order to test consistence of the RAPD amplification process. Problematic bands with no reproducibility across different runs for the same sample were eliminated from further analysis. The same person scored all the RAPD patterns. In order to minimize problematic and spurious bands, all the primers were tested by PCR in isolated mitochondrial DNA with nuclear DNA as positive control. In any case no amplification product was obtained. PCR products were resolved using agarose gel (1.5%) electrophoresis in 1× TAE. Two lines of 100 bp molecular ladder (Roche) were used to estimate molecular weight and to allow for fragment comparison. Gels were stained by immersion in 0.5 μg/ml ethidium bromide for 30 min, visualized on a transilluminator and the image recorded with a UVP (Ultra-Violet Products, Gel Documentation System ImageStore 5000, v7.12) video camera. DNA concentration and molecular weights of the RAPD fragments were estimated using the program Gelworks (Gelwld 2.51) and checked by the same genotype reader. For each sample, the number of bands greater than 100 bp but less than 2,000 bp produced for each sample for each primer was then scored as presence (1) or absence (0) using the molecular ladder as a reference.
Data analysis
The percentage of polymorphic bands (PPB), effective number of alleles (n e), Nei’s (1987) expected heterozygosity (h) and Shannon’s Information Index (I) were obtained with the GENALEX software (Peakall and Smouse 2006) for RAPD data. In order to test if differences in sample size could be responsible for differences in the level of genetic diversity, data was bootstrapped. Ten thousand replicates, with a size equal to the minimum number of individuals analyzed (i.e., 23), were obtained, and the average expected heterozygosity and a 95% confidence interval were computed. The expected heterozygosity was compared among samples with a Friedman test in R (R Development Core Team 2009), with individuals as groups and markers as blocks. A post hoc analysis on the Friedman test was carried out as described in Bortz et al. (2000).
Nei’s (1987) genetic distance, PhiPT (an analogue of the fixation index Fst) and a principal coordinate analysis (PCA) based on a matrix of Euclidean distances (calculated from RAPD marker bands) were computed with GENALEX. PCA is a multivariate analysis used for exploring similarities or dissimilarities in data. The multivariate dataset is presented in a reduced number of dimensions (usually 2 or 3) with each consecutive and uncorrelated axis representing a lower proportion of the original information. A discriminant analysis using the jackknife procedure was performed also in R (R Development Core Team 2009) on the resultant factors of RAPD data, to assess the reliability with which individuals could be ascribed to their respective localities.
The significant differences in marker frequencies among samplings were determined by means of the Markov chain Monte Carlo version of the Fisher’s exact test (R × C test) (Raymond and Rousset 1995; Sokal and Rohlf 1995) implemented in the Tools for Population Genetics Analysis ver. 1.3 software (Miller 1997). Randomization settings included 1,000 dememorization steps, 10 batches, and 2,000 permutations per batch. A global test over loci was calculated using the Fisher’s combined probability test (Sokal and Rohlf 1995).
The phenotypic data of each individual was coded according to the dominant marker model implemented in STRUCTURE 2.3 (Pritchard et al. 2000). This program employs a model-based clustering method to infer population structure and it is useful to determine if the set of pre-defined samples are congruent with genetic information. In order to estimate the number of populations the distribution of the estimated likelihood of K was examined and the ΔK summary statistic of Evanno et al. (2005) was computed. The program was run for under for K values between 1 and 8 under the admixture and non admixture models, considering correlated and independent allele frequencies and employing or avoiding sampling locations as prior information to assist the clustering. For each K value, 10 runs with a 2 × 104 burning followed by a 2 × 104 steps (different runs lengths ranging 104–106 were tested).
Results
The 263 sword razor shells scored for 61 RAPD loci obtained with five primers showed a unique phenotype. The proportion of polymorphic loci was very similar for all samples, ranging from 95.08 to 100%. The effective number of alleles per locus, the Nei’s gene diversity and the Shannon Index (Table 1) were 1.449–1.656, 0.260–0.374 and 0.429–0.551, respectively, with the samples of 2006 presenting the lower values.
The mean expected heterozygosity values obtained after bootstrapping were remarkably similar to those obtained with the original data (the maximum discrepancy between values was 0.018), suggesting that differences in sample sizes could not explain differences between samples (Fig. 2). Additionally, the confidence intervals suggested a clear drop in the expected heterozygosity between Pre-Prestige and 2006 samples. Nevertheless, this change happened differently in Barra and Fisterra. Barra displayed a more stepped change, first between Pre-Prestige and 2004–2005 samples and then between 2004–2005 and 2006 samples. In Fisterra, however, the fall in the expected heterozygosity was mainly due to an abrupt drop in 2006 samples.
Nei’s genetic distance and PhiPT values ranged from 0.046 to 0.244 and from 0.039 to 0.306, respectively (Table 2). Comparisons including either Barra or Fisterra 2006 samples showed in all cases higher values of Nei’s genetic distance and PhiPT (0.189–0.244 and 0.220–0.306, respectively) than comparisons not including one of these samples (0.046–0.176 and 0.039–0.201, respectively).
The Friedman test detected significant differences in the expected heterozygosity among samples (Friedman chi-squared = 26.809, df = 7, p value < 0.001). The post hoc analysis showed four significant comparisons. Of these, two included the pre-Prestige sample of Barra (Barra-pre-Prestige vs. Barra-2006, p value = 0.0038 and Barra-pre-Prestige vs. Fisterra-2006, p value = 0.0086) and the other two ones, the Fisterra-2004 sample (Fisterra-2004 vs. Fisterra-2006. p value = 0.00249, and Fisterra-2004 vs. Barra-2006, p value = 0.0495).
On the 1,708 pairwise exact tests for population differentiation, 218 were significant after sequential Bonferroni correction (Table 3). Comparisons among samples of the same year showed a low-moderate number of significant tests (0–5) and also did those between (2–8) and within (Barra: 7–8, Fisterra: 0–3) sampling sites not including the year 2006 sample. In contrast, comparisons including samples from 2006 showed a considerable higher number of (9–18) significant tests.
Genetic similarities among the 263 E. siliqua individuals for RAPD data were also examined by principal coordinate analysis (PCA). The first three coordinates explained 66.43% of the total variation and the plot of the first two factors supported the previous results, revealing two groupings, one composed by the samples from the year 2006 and another containing the remaining samples (Fig. 3). A discriminant analysis with the two eigenvectors of the PCA indicated that 50.57 and 98.86% of individuals could be assigned to their original sampling and the two groups described, respectively.
Results found for RAPDs using STRUCTURE under different models were congruent and confirmed the clear genetic differentiation observed in the previous analysis. A high value was observed at K = 2 for Evanno et al.’s (2005) ΔK, the other values being close to zero (data not shown). Although the likelihood continued to increase after K = 2, this value seemed to capture the major structure of the data and runs with higher K produced less consistent results.
Discussion
In this work, 61 RAPD loci were employed to analyse the temporal genetic variation and population differentiation of two E. siliqua locations. Samples were taken before and after the Prestige oil spill, a spillage that extensively affected the Galician coasts.
The pre-Prestige samples showed similar levels of genetic diversity, measured as PPB, ne, h and I. These levels were similar to those reported in previous studies for these and other localities of E. siliqua (Fernández-Tajes et al. 2007; Arias et al. 2010) but higher to those obtained in other marine species with similar high larval dispersal potential and sedentary or low mobility adults (e.g., periwinkle Littorina striata, De Wolf et al. 1998; mussel Mytilus galloprovincialis and acorn barnacle Balanus glandula, Li Ma et al. 2000; Pacific oyster Crassostrea gigas, Aranishi and Okimoto 2004; pullet carpet shell Venerupis senegalensis, Joaquim et al. 2010). In the post-Prestige samples of 2004 and 2005, genetic diversity showed similar, although slightly lower, values. On the contrary, a significant reduction in genetic diversity was detected in 2006 when compared with pre-Prestige samples. Additionally, the absence of significant differences in the genetic diversity between samples of the same year and the low levels of genetic differentiation observed for these comparisons (in opposition to those including 2,006 samples) suggests that there is no apparent restriction to gene flow exists between these localities, as has been described in some of the aforementioned studies (Fernández-Tajes et al. 2007; Arias et al. 2010) and in other species (e.g., Mytilus galloprovincialis, Diz and Presa 2009). The reduction in genetic diversity observed in 2006 samplings could be due to different factors such as mutation, selection, migration and genetic drift (or combination of these).
Previous studies in E. siliqua indicate that most localities around the Iberian Peninsula seem to form a single panmictic population, that is, there is a high immigration rate among them (Fernández-Tajes et al. 2007; Arias et al. 2010). This fact, coupled with the high population sizes expected for organisms such as bivalves, should tend to keep the allelic frequencies of population similar. Nevertheless, if populations were to suffer a reduction in size (natural or human-mediated) they would be more sensitive to the effects of genetic drift (see below) and this could lead to changes in the allelic frequencies and random differences among populations.
Genetic drift, understood as changes in the population due to random sampling, is the most plausible explanation for the results observed in this work. Prestige oil compounds are considered as possible carcinogens. Moreover, six of the HAP in Prestige oil and are part of the 16 PAH designated by the United States Environmental Agency as primary contaminants. Therefore, these pollutants might have had lethal effects in E. siliqua individuals, reducing the population size and increasing the effects of genetic drift, which are higher in small populations. Nevertheless, this contrasts with the lack of reports of massive mortalities since an increase in razor clam mortality would have been associated with higher presence of shells in beaches (Fahy et al. 2002) and this has not been documented after oil spillage Alternatively, sub-lethal effects, physiological or ecological, caused by Prestige oil are thought to have occurred in marine organisms exposed to those pollutants or it could also be the case that certain phases of the life cycle (e.g., larvae) might be especially sensitive to pollution. Alterations in physiological processes can result in a fitness decrease (Nony and Schnellmann 2001) and effects of the toxicants on ecological resources such as food accessibility, presence of predators, etc., would indirectly affect survival and/or reproduction of animals in contaminated areas as well (De Wolf et al. 2004b). Sublethal levels of pollutants may thus be associated with the loss of genetic diversity within a population exposed to pollution even though the population as a whole, at least over the short term, is able to survive (Street et al. 1998; Li Ma et al. 2000). In this sense, both the physiologic and the ecological effects may have contributed to a low reproductive success in those individuals affected by the exposure to Prestige oil. Indeed, gametogenesis in E. siliqua specimens starts during November–December and during this period an increase in energy and food supply is needed for gonad development (Darriba et al. 2005). Sub-lethal effects caused by PAH could have implied a deficiency in gonad development and thus a reduced reproductive success in the individuals affected by the Prestige oil spillage. Sublethal effects of exposure to the Prestige fuel (growth reduction and lipid composition) were also reported by Peteiro et al. (2006, 2007) in mussels sampled in 2003. Other sub-lethal effects have also been observed in mussels directly exposed to the Prestige oil, such as DNA damage (Laffon et al. 2006), lysosomal disturbances and alterations of the digestive gland (Marigómez et al. 2006), diminished survival rate in air and changes in lipid and fatty acid composition (Labarta et al. 2005). Therefore, we could expect a bottleneck, with a smaller number of individuals contributing to the next generation or a reduced number of larvae surviving. This is similar to what happens under the “sweepstakes hypothesis” proposed by Hedgecock (1994), under which a small portion of the existing adults would contribute to the next generation, resulting in temporal genetic variation as a function of a stronger genetic drift due to small Ne. A bottleneck is expected to affect most loci, reducing their heterozygosity and polymorphism levels (e.g., Belfiore and Anderson 2001; Theodorakis et al. 2001; De Wolf et al. 2004b) and would cause a loss of the “present bands” in samplings affected by the contamination. The reduction in the expected heterozygosity observed in this work and the levels of genetic differentiation between Pre-Prestige and 2006 samples are congruent with the existence of such a bottleneck.
Heterozygosity values reflect the relationship between the genetic and physiological factors. It is an important criterion for animal fitness since the presence of heterozygous alleles coding for proteins theoretically increase adaptability (Gaffney 1990; Abele et al. 2009). A high degree of heterozygosity at the protein level has also been related to a greater metabolic efficiency, which would allow for higher investments in energy, growth, and reproduction (Koehn and Shumway 1982). Although, there is no correlation between heterozygosity and reproduction and growth in bivalve mobile species, such as razor clams (Beaumont et al. 1985; Bricelj and Krause 1992), a relationship between multi-locus heterozygosity and a higher scope for activity (i.e., swimming and escape responses) has been documented (Alfonsi et al. 1995; Brokordt et al. 2009) Therefore, the decrease in heterozygosity levels observed in E. siliqua would imply a severe loss of alleles and would compromise adaptability of this marine species. Nevertheless, we have to bear in mind that heterozygosity values obtained through dominant markers must be taken in account cautiously because assumptions made with this kind of markers may not be totally realistic since most of the markers analysed, theoretically, should be neutral.
Now, the question is how to explain why the temporal genetic variation was only observed in 2006 samples. Although the establishment of razor clams cohorts is very difficults because it is not possible to determine accurately the age of sampled individuals, we have made an approximation. Studies carried out by Gaspar et al. (1994) and by Da Costa et al. (unpublished data) reported that sizes around 12 cm correspond to 3-year E. siliqua adults. In this work, the size of all specimens was around 12–14 cm; therefore, individuals from 2006 would belong to the offspring spawned after Prestige oil spillage whereas those from 2004 and 2005 would correspond to offspring spawned before the disaster. Thus, the temporal genetic variation described in the current study might indeed be the consequence of a bottleneck provoked by the Prestige oil spillage.
There are some studies on the literature about the effect of Prestige spillage on the genetic population structure of marine invertebrates. Piñeira et al. (2008) studied the genetic impact of the Prestige oil in wild populations of the marine snail Littorina saxatilis one and a half years after the spill. They did not observe a reduction in genetic diversity in exposed populations, concluding that the population either suffered a light bottleneck or had fully recovered from a strong one due to re-colonization processes. Varela et al. (2009) assessed the temporal genetic variation in a natural bed of the razor clam Ensis arcuatus affected by the Prestige oil-spillage. A slight temporal variation was observed in 2003 and 2004 samplings. A high variance in genetic success and/or the movement of adults or larvae from different source populations were the hypothesis proposed by the authors to explain this variation. These results contrast with the high genetic temporal variation observed in this work. Some reasons can be given to explain this observation: (1) the sampling period analysed after Prestige oil spill, which was wider in this work, and (2) the biological characteristics of each species (e.g., dispersal ability, distribution, resistance to pollutants or habitat), which surely have a great influence in how organisms resist pollution events.
The results obtained in this study showed the existence of temporal genetic variation in populations of E. siliqua several years after the Prestige oil spillage. This drop in genetic variability has probably been caused by a strong genetic drift due to a reduction in population size. Nevertheless, and although our work suggests a direct link with the Prestige oil spillage, the natural-mediated variation cannot be excluded as a possible explanation to the observed genetic variation in 2006 samples. A much longer temporal sequence or much broader spatial sampling, including uncontaminated areas and locations with otherwise similar ecological features would offer an additional support to our investigation. Notably, this study reinforces the importance of long time monitoring of contamination events since their effects might not be evident in the directly affected cohorts. This fact is particularly important in long-lived species or species with a long-life larval stage, where the effects of pollutants can appear several years after the disaster had happened. Furthermore, it is necessary to obtain an adequate knowledge of the life history of the species analysed, given that physiological and reproductive traits such as investment in offspring influence how compounds can interact at individual and population level. Finally, our study highlights the suggestion from Ross et al. (2002) that assessment of the genetic diversity of populations could be a valuable addition, as ecological indicator, to more traditional tools for determining the effects of environmental pollution on aquatic ecosystems.
References
Abele D, Brey T, Philipp E (2009) Bivalve models of aging and the determination of molluscan lifespans. J Exp Ger. doi: 10.1016/j.exger.2009.02.012
Alfonsi CY, Nusetti O, Perez J (1995) Heterozygosity and metabolic efficiency in the scallop Euvola ziczac (Linneaus, 1758). J Shellfish Res 14:389–393
Aranishi F, Okimoto T (2004) Genetic relationship between cultured populations of Pacific oyster revealed by RAPD analysis. J Appl Genet 45:435–443
Arias A, Fernández-Moreno M, Fernández-Tajes J, Gaspar MB, Méndez J (2010) Strong genetic differentiation among east Atlantic populations of the sword razor shell (Ensis siliqua) assessed with mtDNA and RAPD markers. Helgol Mar Res. doi: 10.1007/s10152-010-0203-6
Avise JC (2000) Phylogeography, the history and formation of species. Harvard University Press, Cambridge
Beaumont AR, Gosling EM, Beveridge CM, Budd MD, Burnell GM (1985) Studies on heterozygosity and size in the scallop, Pecten maximus. In: Gibbs PE (ed) Proceedings of the 19th European marine biology symposium. Cambridge University Press, Cambridge, pp 443–454
Belfiore NM, Anderson SL (2001) Effects of contaminants on genetic patterns in aquatic organisms: a review. Mutat Res 489:97–122
Bickham JW, Sandhu S, Hebert PDN, Chikhi L, Athwal R (2000) Effects of chemical contaminants on genetic diversity in natural populations: implications for biomonitoring and ecotoxicology. Mutat Res 463:33–51
Bortz J, Lienert G, Boehnke K (2000) Verteilungsfreie Methoden in der Biostatistik. Springer, Berlin
Bricelj VM, Krause MK (1992) Resource allocation and population genetics of the bay scallop, Argopecten irradians irradians: effects of age and allozyme heterozygosity on reproductive output. Mar Biol 113:253–261
Brokordt K, Leiva N, Jeno K, Martínez G, Winkler F (2009) Effect of allozyme heterozygosity on basal and induced levels of heat shock protein (Hsp70), in juvenile Concholepas concholepas (Mollusca). J Exp Mar Biol Ecol 370:18–26
Calderón I, Palacín C, Turon X (2009) Microsatellite markers reveal shallow genetic differentiation between cohorts of the common sea urchin Paracentrotus lividus (Lamarck) in northwest Mediterranean. Mol Ecol 18:3036–3049
Chung PP, Hyne RV, Mann RM, O. Ballard JW (2010) Temporal and geogrpaphical variation in the amphipod Melita plumulosa (Crustacea: Melitidae): link of a localized change in haplotype frequencies to a chemical spill. Chemosphere. doi: 10.1016/j.chemosphere.2010.10.043
Da Costa F, Martínez-Patiño D, Ojea J, Novoa S (2010) Larval rearing and spat production of the razor clam Ensis siliqua (Bivalvia: Pharidae). J Shell Res 29:347–351
Darriba S, San Juan F, Guerra A (2005) Gametogenic cycle of Ensis siliqua (Linnaeus, 1758) in the ría de Corcubión, northwestern Spain. J Moll Stud 71:47–51
De Wolf H, Backeljau T, Verhagen R (1998) Congruence between allozyme and RAPD data in assessing macrogeographical genetic variation in the periwinkle Littorina striata (Mollusca, Gastropoda). Heredity 81:486–492
De Wolf H, Blust R, Backeljau T (2004a) The use of RAPD in ecotoxicology. Mutat Res 566:249–262
De Wolf H, Blust R, Backeljau T (2004b) The population genetic structure of Littorina littorea (Mollusca, Gastropoda) along a pollution gradient in the Scheldt estuary (The Netherlands) using RAPD analysis. Sci Total Environ 325:59–69
Diz AP, Presa P (2009) The genetic diversity pattern of Mytilus galloprovincialis in Galician Rias (NW Iberian estuaries). Aquaculture 287:278–285
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fahy E, Gaffney J (2001) Growth statistics of an exploited razor clam (Ensis siliqua) bed at Gormanstown, Co Meath, Ireland. Hydrobiologia 465:139–151
Fahy E, Alcantara ML, Norman M, Browne R, Roantree V, Pfeiffer N (2002) Mortalities of Ensis arcuatus (Jeffreys) (Solenacea) in western Ireland. J Shell Res 21:29–32
Fernández-Tajes J, Gaspar M, Martínez-Patiño D, McDonough N, Roberts D, González-Tizón A, Martínez-Lage A, Méndez J (2007) Genetic variation of the razor clam Ensis siliqua (Jeffreys 1875) along the European coast based on random amplified polymorphic DNA markers. Aquac Res 38:1205–1212
Figueras A, Lora-Tamayo E, Tintoré J, Pérez F, Albaigés J, Sánchez F, Murado AG, Rodríguez EE, Hernán R (2005) Las lecciones de la catastrophe del Prestige. Consejo superior de Investigaciones Científicas, Madrid
Gaffney PM (1990) Enzyme heterozygosity, growth rate and viability in Mytilus edulis, another look. Evolution 44:204–209
Gaspar MB, Monteiro CC (1998) Reproductive cycles of the razor clam Ensis siliqua and the clam Venus striatula off Vilamoura, Southern Portugal. J Mar Biol Assoc UK 78:1247–1258
Gaspar MB, Richardson CA, Monteiro CC (1994) The effects of dredging on shell formation in the razor clam Ensis siliqua from Barrinha, southern Portugal. J Mar Biol Assoc UK 74:927–938
Hedgecock D (1994) Does variance in reproductive success limit effective population sizes of marine organisms). In: Beaumont AR (ed) Genetics and evolution of aquatic organisms. Chapman and Hall, London, pp 122–134
Joaquim S, Pereira J, Leitao A, Matías D, Chaves R, Guedes-Pinto H, Chícharo L, Gaspar M (2010) Genetic diversity of two Portuguese populations of the pullet carpet shell Venerupis senegalensis, based on RAPD markers: contribution to a sustainable restocking program. Helgol Mar Res 64:289–295
Johnston EL, Roberts DA (2009) Contaminants reduce the richness and evenness of marine communities: a review and meta-analysis. Environ Pollut 157:1745–1752
Koehn RK, Shumway SE (1982) A genetic/physiological explanation for differential growth rate among individuals of the American oyster, Crassostrea virginica (Gmelin). Mar Biol Lett 3:35–42
Körpe DA, Aras S (2011) Evaluation of copper-induced stress on eggplant (Solanum melongena L.) seedlings at the molecular and populations levels by use of various biomarkers. Mutat Res 19:29–34
Labarta U, Fernández-Reiriz MJ, Garrido JL, Babarro JMF, Bayona JM, Albaigés J (2005) Response of mussel recruits to pollution from the Prestige oil spill along the Galicia coast. A biochemical approach. Mar Ecol Prog Ser 302:135–145
Laffon B, Rábade T, Pásaro E, Méndez J (2006) Monitoring of the impact of Prestige oil spill on Mytilus galloprovincialis from Galician coast. Environ Int 32:342–348
Li Ma X, Cowles DL, Carter RL (2000) Effect of pollution on genetic diversity in the bay mussel Mytilus galloprovincialis and the acorn barnacle Balanus glandul. Mar Environ Res 50:559–563
Liu W, Yang YS, Zhou Q, Xie L, Li P, Sun T (2007) Impact assessment of cadmium contamination on rice (Oryza sativa L.) seedlings at molecular and population levels using multiple biomarkers. Chemosphere 67:1155–1163
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Marigómez I, Soto M, Cancio I, Orbea A, Garmendia L, Cajaraville MP (2006) Cell and tissue biomarkers in mussel, and histopathology in hake and anchovy from Bay of Biscay after the Prestige oil spill (Monitoring Campaign 2003). Mar Pollut Bull 53:287–304
Matson CW, Lambert MM, McDonald TJ, Autenrieth RL, Donnelly KC, Islamzadeh A, Politov DL, Bickham JW (2006) Evolutionary toxicology: population-level effects of chronic contaminant exposure on the marsh frogs (Rana ridibunda) of Azerbaijan. Environ Health Perspect 114:547–552
Miller MP (1997) Tools for Population Genetic Analyses (TFPGA) 1.3: a Windows Program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author
Monteiro C, Gaspar M (1993) Bivalves do litoral oceânico algarvio: breve noticia sobre a situaçâo actual dos Principais bancos (Julho 1993). In: Relatório Tecnico Cientifico INIP, Lisboa, 65, p 19
Nadig SG, Lee KL, Adams SM (1998) Evaluating alterations of genetic diversity in sunfish populations exposed to contaminants using RAPD assay. Aquat Toxicol 43:163–178
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nony PA, Schnellmann RG (2001) Interactions between collagen IV and collagen-binding integrins in renal repair after sublethal injury. Mol Pharmacol 60:1226–1234
Palumbi SR (2004) Marine reserves and ocean neighbourhoods: the spatial scale of marine populations and their management. Ann Rev Environ Res 29:31–68
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peteiro LG, Bábarro JMF, Labarta U, Fernández-Reiriz MJ (2006) Growth of Mytilus galloprovincialis after the Prestige oil spill. ICES J Mar Sci 63:1005–1013
Peteiro LG, Labarta U, Fernández-Reiriz MJ (2007) Variability in biochemical components of the mussel (Mytilus galloprovincialis) cultured after Prestige oil spill. Comp Biochem Physiol C 145:588–594
Piñeira J, Quesada H, Rolán-Alvarez E, Caballero A (2008) Genetic impact of the Prestige oil spill in wild populations of a poor disperse marine snail from intertidal rocky shores. Mar Poll Bull 56:270–281
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Ross K, Cooper N, Bidwell JR, Elder J (2002) Genetic diversity and metal tolerance of two marine species: a comparison between populations from contaminated and reference sites. Mar Pollut Bull 44:671–679
Sokal RR, Rohlf FL (1995) Biometry, 3rd edn. WH Freeman and Company, New York
Street GT, Lotufo GR, Montagna PA, Fleeger JW (1998) Reduced genetic diversity in a meiobenthic copepod exposed to a xenobiotic. J Exp Mar Biol Ecol 222:93–111
R Development Core Team (2009) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0. http://www.R-project.org
Theodorakis CW, Bickham JW (2004) Molecular characterization of contaminant-indicative RAPD markers. Ecotoxicology 13:303–309
Theodorakis CW, Bickham JW, Lamb T (2001) Integration of genotoxicity and population genetic analyses in kangaroo rats (Dipodomys merriami) exposed to radionuclide contamination at the Nevada test site, USA. Environ Toxicol Chem 20:317–326
Varela MA, Martínez-Lage A, Gonzalez-Tizon AM (2009) Temporal genetic variation of microsatellite markers in the razor clam Ensis arcuatus (Bivalvia: Pharidae). J Mar Biol Assoc UK 89:1703–1707
Von Cosel R (1990) An introduction to the razor shells (Bivalvia: Solenacea). In: Morton B (ed) The Bivalvia proceedings of a memorial symposium in honour of Sir Charles Maurice Yongue. Hong Kong University Press, Hong Kong, pp 283–305
Acknowledgments
This work was funded by a PGIDIT03 RMA10301PR, and PGIDIT03 PX10302PR grants from Galicia Government and by PFSHARE and TIMES grants from Community Initiative Programme INTERREG-IIIB “Atlantic Area”. We are grateful to Mr. Jose García Gil for his technical assistance and to two anonymous reviewers for their valuable comments in the text.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Fernández-Tajes and A. Arias-Pérez have equally contributed to this work.
Rights and permissions
About this article
Cite this article
Fernández-Tajes, J., Arias-Pérez, A., Fernández-Moreno, M. et al. Sharp decrease of genetic variation in two Spanish localities of razor clam Ensis siliqua: natural fluctuation or Prestige oil spill effects?. Ecotoxicology 21, 225–233 (2012). https://doi.org/10.1007/s10646-011-0781-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-011-0781-3