Abstract
Elemental diet (ED) has been used as an enteral nutritional therapy for Crohn’s disease. However, the precise mechanisms of ED remain unclear. In interleukin-10 (IL-10)-deficient cell-transferred mice, we investigated the change of intestinal microbiota with ED using molecular terminal-restriction fragment length polymorphism (T-RFLP) analysis and culture method, and evaluated its influence on therapeutic effects of ED. ED significantly suppressed intestinal inflammation. The total amount of bacteria in colitis mice fed the regular diet was higher than in normal mice but decreased in colitis mice fed ED. T-RFLP profiles of the ED group markedly differed from those of the regular diet groups. The diversity of bacterial species in the ED group decreased to 60% of that found in the regular diet groups. Among the cultivated bacteria, the change in lactic acid bacteria composition was remarkable. Lactobacillus reuteri and L. johnsonii decreased and Enterococcus faecalis and E. durans increased in the ED group. The culture supernatant of L. reuteri isolates induced significant tumor necrosis factor-alpha (TNF-α) and IL-6 activity in RAW 264 cells, while the culture supernatant of E. faecalis and E. durans barely induced their activity. These data suggested that reduction in amount and diversity of intestinal microbiota and decrease of proinflammatory cytokines via a change in composition of lactic acid bacteria by ED seem to contribute to reduction of bowel inflammation in this model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammatory bowel disease (IBD) is supposed to be the result of interactions among genetic, immune, and environmental factors [1]. Intestinal microbiota is an important environmental factor in the etiology of IBD. Antibodies against Saccharomyces cerevisiae (ASCA), Pseudomonas fluorescens-associated sequence I2 and the outer membrane porin protein C of Escherichia coli (OmpC) have been specifically detected in patients with Crohn’s disease (CD) [2–6]. On the other hand, no inflammation or colitis is induced in certain murine and rodent models maintained under germ-free conditions [7, 8]. Among animal models, interleukin-10-deficient (IL-10−/−) mice have been used to investigate the influence of microbiota on the development of colitis, because the effect of intestinal microbiota on the immune response to intestinal inflammation is associated with an apparently dysregulated production of Th1-type proinflammatory cytokines, similar to that observed in CD [9–12]. Some modifications of the enteric microbiota can attenuate colitis in this model [13, 14].
In Japan, elemental diet (ED; Elental®, Ajinomoto Co. Inc., Tokyo, Japan) has been used as the primary therapy for CD, showing an effectiveness equivalent to that of corticosteroids [15–18], but the precise mechanisms underlying the therapeutic effects of ED remain unclear. ED is a special formula that contains amino acids as the source of elemental nitrogen, together with other easily digestible nutrients, minerals, and vitamins. Fat is present in a very small quantity. The intestinal microbiota is strongly influenced by nutrients; thus the intestinal microbiota of CD patients treated with ED is assumed to change.
The objectives of this study were: (i) to examine the effect of ED using IL-10−/− cell-transferred mice showing chronic inflammations and immunological characteristics with human CD [19], (ii) to characterize the differences in the intestinal microbiota between mice fed a regular diet and those fed the elemental diet, and (iii) to elucidate whether the change in intestinal microbiota contributed to the therapeutic effects of ED.
Materials and Methods
Animal Experiment
IL-10−/− cell-transferred colitis mice were used. Transfer of IL-10−/− cells was done using the methods described by Ikenoue et al. [19]. Briefly, spleen and mesenteric lymph node (MLN) cells from diseased male IL-10−/− mice were isolated and single-cell suspensions were prepared by mechanical dissociation. Erythrocytes were removed by hypotonic lysis. Female CB-17 SCID mice, 8–12 weeks of age, were injected intraperitoneally 1.0 × 107 cells suspended in 200 μl phosphate buffered saline (PBS). IL-10−/− cell-transferred and nontransferred mice were fed a regular diet (CRF-1, Charles River Japan, Inc., Tokyo, Japan) or elemental diet (dextrin 79.3%, amino acids 17.6%, soybean oil 0.6%, minerals 2.0%, vitamins 0.5%, by weight) for 1, 2, and 3 weeks. Three groups of mice were used: cell-transferred mice fed the regular diet (TRD group); cell-transferred mice fed the elemental diet (TED group); and nontransferred mice fed the regular diet (NRD group) as the healthy control. The mice were sacrificed, then their cecum and colon were removed and weighed. Cecal contents were used for the analysis of microbiota and short-chain fatty acids (SCFAs) while the colon was histologically examined. Each sample was identified by group name, breeding period, and mouse identification (ID) number.
Histological Analysis
Tissues samples were fixed in 10% phosphate-buffered formalin. Paraffin-embedded sections were stained with hematoxylin and eosin (H–E) for light microscopic examination.
Cell Lysis and DNA Isolation from Cecal Samples
DNA was isolated using Ultra CleanTM Soil DNA Isolation Kit (Mo Bio Laboratories Inc., Solana Beach, CA). The sample (0.05 g) was suspended in a bead solution containing 5 mg/ml lysozyme and 0.08 mg/ml N-acetylmuramidase, and incubated for 30 min at 37°C to lyse the cells. DNA was extracted and purified as described by Clement and Kitts [20] with some modifications.
PCR and T-RFLP Analysis
Polymerase chain reaction (PCR) amplification was performed as described by Kibe et al. [21]. Two pairs of primers were used: 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) as the universal primer, and 27f and Lab-677r (5′-CACCGCTACACATGGAG-3′) as the specific primer set for lactic acid bacteria (LAB) designed by Heilig et al. [22]. 27f was labeled with 6-FAM (6-carboxyfluorescein, Applied Biosystems, Foster City, CA) for T-RFLP analysis. PCR was performed using a Thermocycler T Gradient (Biometra, Gottingen, Germany) in a reaction mixture (50 μl) containing 5 μl dissolved DNA (100 ng), 1.25 U TaKaRa Ex TaqTM (Takara, Shiga, Japan), 10 × Ex TaqTM buffer, 4 μl dNTP mixture (2.5 mM each), and 10 pmol of each primer. The amplification program used was as follows: preheating at 95°C for 3 min; 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1.5 min; and final extension at 72°C for 10 min. The PCR products were purified by polyethylene glycol (PEG) precipitation method [23]. Purified PCR products were digested with 20 U of either HhaI or MspI (Takara) in total volume of 10 μl at 37°C for 3 h. Length of the terminal-restriction fragments (T-RFs) was determined based on standard size markers GS500 ROX and 1000 ROX (Applied Biosystems) using the ABI PRISMTM 3100 genetic analyzer (Applied Biosystems) and GeneScan® analysis software (Applied Biosystems). A dendrogram analysis was performed using T-RFLP patterns by using BioNumerics software (Applied Maths Sint-Martens-Latem, Belgium). Distances between samples were represented graphically by constructing a dendrogram based on Jaccard’s similarity coefficients and Ward’s algorithm type. The position tolerance of band matching was 0.05%.
Analysis of Short-Chain Fatty Acids
A portion of cecal contents was deproteinated with perchloric acid and used to measure short-chain fatty acids (SCFAs). Cecal SCFA content was measured as follows: the sample was centrifuged and the supernatant was subjected to high-performance liquid chromatography as described by Teramoto et al. [24].
Culture and Isolation of Bacteria
Culturable bacteria in the sample obtained at the end of the experiment (week 3) were detected and isolated by the serial dilution method. Fresh cecal contents were serially diluted with prereduced dilution buffers, and 0.05 ml samples of 106, 107, and 108 dilutions were plated on nonselective BL agar (Nissui Seiyaku, Tokyo, Japan), EG agar (Merck, Darmstadt, Germany), and TS agar (Becton Dickinson, Franklin Lake, NJ); and 0.05 ml of 101,103, 105, and 107 dilutions were plated on selective LBS agar (Becton Dickinson) for Lactobacillus, and DHL agar (Nissui Seiyaku) for Enterobacteria [25]. BL, EG, and LBS agar plates were subsequently incubated at 37°C for 2 days under anaerobic conditions with CO2 gas, while TS and DHL agar plates were subsequently incubated at 37°C for 1 day under aerobic conditions. Bacteria were identified by macroscopic observation of the colonies and microscopic examination of cells with Gram staining.
Isolation and Identification of Lactic Acid Bacteria (LAB)
Twenty-four strains of Lactobacillus growing on LBS agar plates and 16 strains found on TS agar plates and presumed to be Streptococcus or Enterococcus, were transferred with a sterile toothpick to 50 μl 10 mM Tris–HCl-1 mM ethylenediamine tetraacetic acid (EDTA) (pH 8.0) (TE buffer). One microliter of the suspension was smeared onto a glass slide for Gram staining. The remaining suspension was boiled for 15 min to lyse the cells and the lysate was used as template for the PCR. Nearly complete (1,500 bases) 16 S rRNA gene sequences of the strains investigated were amplified by PCR using universal primers 27f and 1492r. PCR products were sequenced using a BigDye Terminator cycle sequencing kit and the ABI PRISMTM 3100 Genetic Analyzer (both from Applied Biosystems). The closest relatives of the isolates were identified through database searches, and the sequences of closely related species were retrieved from DNA Data Bank of Japan (DDBJ), Europian Molecular Biology Laboratory (EMBL), and Genbank.
Cytokines Induction of LAB Culture Supernatant in the Murine Macrophage Cell Line
Isolates identified as LAB were cultured in Lactobacillus MRS broth (Becton Dickinson) for 2 days at 37°C under anaerobic conditions. In vitro cytokines induction assays were carried out using LAB culture supernatant as described by Peña et al. [26] with some modifications. Briefly, media conditioned by LABs were tested for the ability to induce the production of TNF-α and IL-6 by the murine macrophage cell line RAW 264 (JCM RCB0535). RAW 264 macrophages were incubated in Dulbecco’s Modified Eagles Medium (D-MEM; Sigma-Aldrich, Taufkirchen, Germany) supplemented with 10% fetal bovine serum (FBS; MP Biomedicals LLC, Morgan Irvine, CA), 20 units/ml sodium penicillinG, and 20 μg/ml streptomycin sulfate for 3 days and then exposed to 5% (v/v) LAB culture supernatant in 24-well flat-bottom plates. After 6 h, the culture supernatant was collected and TNF-α and IL-6 production was measured using a BD Opt EIATM Mouse TNF Mono/Mono Set (Becton Dickinson) and a BD Opt EIATM Mouse IL-6 enzyme-linked immunosorbent assay (ELISA) Set (Becton Dickinson), respectively.
Statistical Analysis
Data are presented as mean ± standard error (SE). Differences between two groups were evaluated using Student’s t-test and Dunnett’s test. The level of statistical significance was set at <0.05.
Results
Effect of ED on Inflammation in Colitis
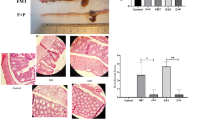
We assessed the therapeutic effects of ED in IL-10 (−/−) cell-transferred mice. As shown in Fig. 1a, mice in the TRD group progressively lost weight during the 3 weeks after cell transfer. In contrast, mice in the TED group did not lose weight. At week 1, colon weight was similar in the three groups, but at weeks 2 and 3 it increased significantly due to inflammation in the TRD group, while in the TED group it was significantly lower than in the TRD group (Fig. 1b). At week 3, histological examination of the colon revealed massive inflammatory infiltrates, crypt hyperplasia, and marked mucin depletion of goblet cells in the TRD group, whereas in the TED group histological damage was markedly suppressed (Fig. 1c, H–E staining).
Weight and SCFAs Content in the Cecum
Cecal weight and SCFAs content measured at week 3 after cell transfer are shown in Table 1. Cecal weight in the TRD3W group was significantly greater than that in the NRD3W group, and in the TED3W group it was significantly smaller than in the TRD3W group (P < 0.01). We also measured cecal SCFAs content. Total SCFA concentration in the TRD3W group was about twofold that in the NRD3W group. Concentrations of malic acid, succinic acid, and lactic acid were especially higher in the TRD3W group compared with in the other two groups. Higher concentrations of propionic acid and valeric acid were characteristic of the TED3W group.
T-RFLP
Remarkable differences in T-RFLP profiles were observed between the TED group and the other two groups, while the difference between NRD and TRD groups was quite small (Fig. 2). In the TED group, many T-RFs disappeared, the proportion of these T-RFs decreased, and a few new T-RFs emerged or the proportion of specific T-RFs increased. A cluster analysis done by combining T-RFLP profiles derived from MspI and HhaI revealed that all samples from the TED group distributed within its cluster while most samples from the NRD and TRD groups were in the same cluster (Fig. 3). The difference between the TED group and NRD group or TRD group was more marked than between the NRD group and TRD group. The similarity index increased in relation to the breeding period. The number of T-RFs in the TED group decreased to 60% of that in the other groups (Fig. 4). The alteration in the composition of LAB in the TED group was also confirmed by T-RFLP profiles. The size of some T-RFs obtained with universal and LAB primers coincided, indicating that LAB accounted for a large proportion of the bacterial population in this murine intestinal microbiota.
T-RFLP pattern of 16 S rDNA from the cecum digested with HhaI (left) and MspI (right). a 16 S rDNA was amplified with universal primers. b LAB specific primers. Arrows indicate the Lactobacillus spp. determined from the DNA sequence data of the isolates listed in Table 3
Cultivation of Predominant Bacteria
The population of major groups of cecum-derived cultivated bacteria is shown in Table 2. Among anaerobic bacteria, the genera Bacteroides and Clostridium predominated, and their population did not differ among the three groups. On the other hand, the population of Enterobacteria and LAB differed among the three groups. Especially, the difference in LAB composition, including the genera Lactobacillus, Streptococcus, and Enterococcus, was significantly distinct. The population of Lactobacillus in the cecum was approximately 2 × 109 cells/g in the NRD3W and TRD3W groups and approximately 1 × 107 cells/g in the TED3W group, whereas the genus Enterococcus was detected only in the TED3W group.
Isolation and Determination of LAB Species
LAB species from colonies isolated on agar plates were identified to clarify the change in LAB (Tables 3 and 4). In TRD3W mice, most Lactobacillus isolates were identified as Lactobacillus reuteri by 16 S rRNA gene sequence and three isolates were identified as L. johnsonii. In the TED3W mice, most of the Lactobacillus isolates were identified as L. murinus. As for the genera Streptococcus and Enterococcus, most isolates in the TED3W group were identified as Enterococcus faecalis and E. durans.
Cytokines Induction in RAW264 Cells by LAB Culture Supernatant
The culture supernatant of all L. reuteri isolates obtained from mice fed the regular diet significantly induced TNF-α (14,200 ± 2,300 pg/ml, P < 0.001) and IL-6 (643 ± 91 pg/ml, P < 0.001), whereas the culture supernatant of L. murinus isolates obtained from the TED3W group induced half the activity of TNF-α and a quarter of the activity of IL-6 compared with L. reuteri (Fig. 5). On the other hand, the culture supernatants of E. faecalis and E. durans isolates obtained from the TED3W group did not induce a significant production of TNF-α, and IL-6 was not detected at all.
Discussion
ED therapy has been established as a primary treatment for CD patients in Japan and pediatric CD patients in Western countries. These effects are reported by some randomized controlled trials [27–30]. However, the therapeutic mechanisms have not been fully clarified.
We used IL-10−/− cell-transferred mice [19] to elucidate the effect of ED treatment. This model shows severe symptoms in short term with a simple procedure that is easy to manipulate and also shows the same Th-1-predominant immune abnormality as IL-10−/− mice. In this study, cell-transfer-induced colonic inflammation and colon weight increased according to severity of inflammation. ED significantly inhibited colonic inflammation macroscopically and microscopically (Fig. 1). In addition, it was observed that plasma level of serum amyloid A (SAA), which is elevated in case of systemic inflammation, significantly decreased in the TED group (data not shown).
We speculated that modifications of the intestinal microbiota in mice fed the ED might be associated with inhibition of colitis in this model. Therefore, we examined the changes in the intestinal microbiota of these mice.
The role of the intestinal microbiota in the pathogenesis of CD has been mainly investigated in cultured bacteria derived from feces. However, no conclusions have been obtained so far because of the heterogeneity and complexity of the human gut microbiota.
In this model, we could control the diet and examine its effect on the intestinal microbiota. Figure 3 shows close similarity of the microbiota among the three mice of each group, indicating that this model is appropriate for analysis of the intestinal microbiota.
Cecal weight in the TRD3W group that exhibited severe colitis was significantly greater than that in the NRD3W group. Additionally, total SCFAs concentration in the TRD3W group was about twice the concentration in the NRD3W group. These findings suggested that the total amount of bacteria was increased in colitis mice. The reason is unknown, although it was suspected that antibacterial properties such as defensins might be decreased in this model, as reported in CD patients and colitis mice [31, 32]. The high ratio of succinic acid and lactic acid observed in the TRD3W group may aggravate colitis [33, 34]. On the other hand, cecal weight in the TED3W group was significantly lighter than that in the NRD3W and TRD3W groups. The cecum contains undigested components and their fermented bacteria. It is well known that ED is fully absorbed in the ileum and that the amount of undigested components is significantly reduced. Therefore, reductions of total amount of intestinal bacteria and its metabolites were strongly expected. Reductions of fecal microbiota and SCFA concentration using standard enteral formula were reported in healthy subjects [35–37]. On the other hand, the change of intestinal microbiota by enteral nutrition has not been sufficiently investigated in clinical studies of IBD and inflammatory bowel syndrome (IBS). In pediatric CD patients, profound modification of fecal microbiota was observed after exclusive enteral nutrition for 8 weeks with a polymeric formula (Modulen IBD®, Nestlé) [38]. Elemental diet (Vivonex Plus® Novaritis Nutrition Corp. Minneapolis, MN) proved to be highly effective in normalizing abnormal lactulose breath caused by bacterial overgrowth in IBS patients [39].
Total concentration of SCFAs in the TED 3 week group was low compared with in the TRD3W group and similar to that in the NRD3W group, but the profiles of SCFAs differed. High ratio of acetic acid and propionic acid, recently reported to have anti-inflammatory effects [40], was found in the TED3W group.
To further investigate the alteration of microbiota by ED, we used the T-RFLP method [41]. This method is relatively simple, easy, and reproducible compared with culture methods [42]. It is also advantageous that T-RFs are indicative of the genus or species of bacteria based on differences in 16 S rRNA sequence of each bacterium and allows the search of candidates genus or species from databases. In this study, T-RFLP profiles of the TED group differed markedly from those of the NRD and TRD groups. The diversity of intestinal microbiota in the TED group was 60% of that found in the NRD and TRD groups (Fig. 4). Some of the T-RFs that disappeared in the TED group were identified as Lactobacillus spp. by LAB specific primers but the others were unidentified. We also examined the gut microbiota using the culture method. We found that the genera Bacteroides and Clostridium predominated and that the population of these anaerobes was similar in the three groups while the population of Enterobacteria and LAB differed. As for the change of LAB, a decrease of L. reuteri and L. johnsonii and an increase of E. faecalis and E. durans were observed in the TED group.
LAB including the genera Lactobacillus and Enterococcus are used as probiotics. Probiotics exert various immunoactivator or immunomodulator effects. Some Lactobacillus strains have been tested for treatment or prevention of recurrence for IBD [8, 43, 44]. It is reported [24] that in IL-10−/− mice Lactobacillus spp. differ from those found in other gene-deficient mice and that they stimulate the murine macrophage cell line RAW 264.7 in different ways. In IBD, macrophages are the primary source of TNF-α that is currently the main target of immunotherapy for CD. Therefore, we investigated the cytokine-promotion activity of LAB isolates using the murine macrophage cell line RAW 264 (Fig. 5). The culture supernatant of L. reuteri recovered from the TRD groups induced production of TNF-α and IL-6, while that of E. faecalis and E. durans isolated from the TED group did not, suggesting that some resident Lactobacillus spp. may be a risk factor for inflammation and that modification of the composition of LAB population by ED contributed to the reduction of inflammation.
It is strongly suggested that loss of tolerance towards resident intestinal microbiota occurs more commonly in animal IBD models than in humans [45]. In this model, transfer of lymphocytes from diseased IL-10−/− mice provoked an inflammatory reaction against the resident intestinal microbiota in SCID mice. In the CD4 + CD45RB high T cell transfer mice [46], colitis developed under specific pathogen-free (SPF) conditions plus segmented filamentous bacteria (SFB), but not under germ-free conditions. Investigation of SFB in this model will be needed in the future. In a T cell receptor alpha-chain-deficient model, Kishi et al. [47] reported that mice fed ED showed no pathologic features of IBD and that pathogenic Bacteroides vulgatus were eliminated by ED.
As mentioned above, various microbiological changes were observed after ED feeding in this IL-10−/− transfer mice: (i) reduction of population and diversity of the microbiota, (ii) change of SCFAs concentration and profile, and (iii) modification of the composition of LAB population that led to a reduction of inflammatory cytokines in the macrophage cell line in vitro. Modification of the microbiota by ED may be responsible, at least in part, for the suppression of the inflammatory response in this model. We expect to confirm these changes of the intestinal microbiota in patients with Crohn’s disease treated with ED.
References
Isaacs KL, Lewis JD, Sandborn WJ, Sands BE, Targan SR. State of the art: IBD therapy and clinical trials in IBD. Inflamm Bowel Dis. 2005;11(l):S3–S12.
Landers CJ, Cohavy O, Misra R, et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto-and microbial antigens. Gastroenterology. 2002;123:689–699. doi:10.1053/gast.2002.35379.
Main J, McKenzie H, Yeaman GR, et al. Antibody to Saccharomyces cerevisiae (bakers’ yeast) in Crohn’s disease. BMJ. 1988;297:1105–1106.
Mow WS, Vasiliauskas EA, Lin YC, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–424. doi:10.1053/j.gastro.2003.11.015.
Quinton JF, Sendid B, Reumaux D, et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut. 1998;42:788–791.
Sutton CL, Kim J, Yamane A, et al. Identification of a novel bacterial sequence associated with Crohn’s disease. Gastroenterology. 2000;119:23–31. doi:10.1053/gast.2000.8519.
Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, Weaver CT. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi:10.1111/j.0105-2896.2005.00291.x.
Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–1633. doi:10.1053/j.gastro.2004.03.024.
Davidson NJ, Leach MW, Fort MM, et al. T helper cell 1-type CD4+ T cells, but not B cells, mediate colitis in interleukin 10-deficient mice. J Exp Med. 1996;184:241–251. doi:10.1084/jem.184.1.241.
Fox JG, Gorelick PL, Kullberg MC, Ge Z, Dewhirst FE, Ward JM. A novel urease-negative Helicobacter species associated with colitis and typhlitis in IL-10-deficient mice. Infect Immun. 1999;67:1757–1762.
Kullberg MC, Jankovic D, Gorelick PL, et al. Bacteria-triggered CD4(+) T regulatory cells suppress Helicobacter hepaticus-induced colitis. J Exp Med. 2002;196:505–515. doi:10.1084/jem.20020556.
Kullberg MC, Andersen JF, Gorelick PL, et al. Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc Natl Acad Sci USA. 2003;100:15830–15835. doi:10.1073/pnas.2534546100.
Madsen KL, Doyle JS, Tavernini MM, Jewell LD, Rennie RP, Fedorak RN. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology. 2000;118:1094–1105. doi:10.1016/S0016-5085(00)70362-3.
Sydora BC, Tavernini MM, Doyle JS, Fedorak RN. Association with selected bacteria does not cause enterocolitis in IL-10 gene-deficient mice despite a systemic immune response. Dig Dis Sci. 2005;50:905–913. doi:10.1007/s10620-005-2663-0.
O’Moráin C, Segal AW, Levi AJ. Elemental diet as primary treatment of acute Crohn’s disease: a controlled trial. Br Med J (Clin Res Ed). 1984;288:1859–1862.
Okada M, Yao T, Yamamoto T, et al. Controlled trial comparing an elemental diet with prednisolone in the treatment of active Crohn’s disease. Hepatogastroenterology. 1990;37:72–80.
Gorard DA, Hunt JB, Payne-James JJ, et al. Initial response and subsequent course of Crohn’s disease treated with elemental diet or prednisolone. Gut. 1993;34:1198–1202. doi:10.1136/gut.34.9.1198.
Yamamoto T, Nakahigashi M, Umegae S, Kitagawa T, Matsumoto K. Impact of elemental diet on mucosal inflammation in patients with active Crohn’s disease: cytokine production and endoscopic and histological findings. Inflamm Bowel Dis. 2005;11:580–588. doi:10.1097/01.MIB.0000161307.58327.96.
Ikenoue Y, Tagami T, Murata M. Development and validation of a novel IL-10 deficient cell transfer model for colitis. Int Immunopharmacol. 2005;5:993–1006. doi:10.1016/j.intimp.2005.01.009.
Clement BG, Kitts CL. Isolating PCR-quality DNA from human feces with a soil DNA kit. Biotechniques. 2000;28:640–646.
Kibe R, Sakamoto M, Hayashi H, Yokota H, Benno Y. Maturation of the murine cecal microbiota as revealed by terminal restriction fragment length polymorphism and 16S rRNA gene clone libraries. FEMS Microbiol Lett. 2004;235:139–146. doi:10.1111/j.1574-6968.2004.tb09578.x.
Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol. 2002;68:114–123. doi:10.1128/AEM.68.1.114-123.2002.
Sakamoto M, Hayashi H, Benno Y. Terminal restriction fragment length polymorphism analysis for human fecal microbiota and its application for analysis of complex bifidobacterial communities. Microbiol Immunol. 2003;47:133–142.
Teramoto F, Rokutan K, Kawakami Y, et al. Effect of 4G-β-D-galactosylsucrose (lactosucrose) on faecal microflora in patients with chronic inflammatory bowel disease. J Gastroenterol. 1996;31:33–39. doi:10.1007/BF01211184.
Benno Y, Sawada K, Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28:975–986.
Peña JA, Li SY, Wilson PH, Thibodeau SA, Szary AJ, Versalovic J. Genotypic and phenotypic studies of murine intestinal lactobacilli: species differences in mice with and without colitis. Appl Environ Microbiol. 2004;70:558–568. doi:10.1128/AEM.70.1.558-568.2004.
Bamba T, Shimoyama T, Sasaki M, et al. Dietary fat attenuates the benefits of an elemental diet in active Crohn’s disease: a randomized, controlled trial. Eur J Gastroenterol Hepatol. 2003;15:151–157. doi:10.1097/00042737-200302000-00008.
Takagi S, Utsunomiya K, Kuriyama S, et al. Effectiveness of an ‘half elemental diet’ as maintenance therapy for Crohn’s disease: a randomized-controlled trial. Aliment Pharmacol Ther. 2006;24:1333–1340. doi:10.1111/j.1365-2036.2006.03120.x.
Johnson T, Macdonald S, Hill SM, Thomas A, Murphy MS. Treatment of active Crohn’s disease in children using partial enteral nutrition with liquid formula: a randomised controlled trial. Gut. 2006;55:356–361. doi:10.1136/gut.2004.062554.
Borrelli O, Cordischi L, Cirulli M, et al. Polymeric diet alone versus corticosteroids in the treatment of active pediatric Crohn’s disease: a randomized controlled open-label trial. Clin Gastroenterol Hepatol. 2006;4:744–753. doi:10.1016/j.cgh.2006.03.010.
Wehkamp J, Salzman NH, Porter E, et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi:10.1073/pnas.0505256102.
Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi:10.1126/science.1104911.
Vernia P, Caprilli R, Latella G, Barbetti F, Magliocca FM, Cittadini M. Fecal lactate and ulcerative colitis. Gastroenterology. 1988;95:1564–1568.
Hove H, Mortensen PB. Influence of intestinal inflammation (IBD) and small and large bowel length on fecal short-chain fatty acids and lactate. Dig Dis Sci. 1995;40:33–39. doi:10.1007/BF02063938.
Whelan K, Judd PA, Preedy VR, Taylor MA. Enteral feeding: the effect on faecal output, the faecal microflora and SCFA concentrations. Proc Nutr Soc. 2004;165:105–113.
Whelan K, Judd PA, Preedy VR, Simmering R, Jann A, Taylor MA. Fructooligosaccharides and fiber partially prevent the alterations in fecal microbiota and short-chain fatty acid concentrations caused by standard enteral formula in healthy humans. J Nutr. 2005;135:1896–1902.
Winitz M, Adams RF, Seedman DA, Davis PN, Jayko LG, Hamilton JA. Studies in metabolic nutrition employing chemically defined diets. II. Effects on gut microflora populations. Am J Clin Nutr. 1970;23:546–559.
Lionetti P, Callegari ML, Ferrari S, et al. Enteral nutrition and microflora in pediatric Crohn’s disease. J Parenter Enter Nutr. 2005;29:S173–S175. doi:10.1177/01486071050290S4S173.
Pimentel M, Constantino T, Kong Y, Bajwa M, Rezaei A, Park S. A 14-day elemental diet is highly effective in normalizing the lactulose breath test. Dig Dis Sci. 2004;49:73–77. doi:10.1023/B:DDAS.0000011605.43979.e1.
Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;28:2826–2832.
Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522.
Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J Med Microbiol. 2003;52:79–89. doi:10.1099/jmm.0.04991-0.
Fiocchi C. Inflammatory bowel disease pathogenesis: therapeutic implications. Chin J Dig Dis. 2005;6:6–9. doi:10.1111/j.1443-9573.2005.00191.x.
Karimi O, Pena AS, van Bodegraven AA. Probiotics (VSL#3) in arthralgia in patients with ulcerative colitis and Crohn’s disease: a pilot study. Drugs Today (Barc). 2005;41:453–459. doi:10.1358/dot.2005.41.7.917341.
Kim SC, Tonkonogy SL, Albright CA, et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi:10.1053/j.gastro.2005.02.009.
Stepankova R, Powrie F, Kofronova O, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RB(high) CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi:10.1002/ibd.20221.
Kishi D, Takahashi I, Kai Y, et al. Alteration of V beta usage and cytokine production of CD4+ TCR beta beta homodimer T cells by elimination of Bacteroides vulgatus prevents colitis in TCR alpha-chain-deficient mice. J Immunol. 2000;165:5891–5899.
Acknowledgment
The authors would like to thank Dr. Ryoko Kibe for her valuable help with T-RFLP analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kajiura, T., Takeda, T., Sakata, S. et al. Change of Intestinal Microbiota with Elemental Diet and Its Impact on Therapeutic Effects in a Murine Model of Chronic Colitis. Dig Dis Sci 54, 1892–1900 (2009). https://doi.org/10.1007/s10620-008-0574-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-008-0574-6