Abstract
In Slovenia, the unique watershed naturally hosting the marble trout is the Soča River, called Isonzo in Italy. In 1993–1996 molecular data established the existence of extensive hybridization with stocked Atlantic domestic lineages which is a threat for this taxon and for the economy of the country established on the angling tourism. Different management actions have been developed for restoring marble genes since 1996: banning stocking of brown trout, revising fishing regulations for anglers and testing genetically brood stock in hatchery for stocking phenotypic and pure marble fry. This long fight against hybridization was genetically surveyed using allozymes, mitochondrial sequences and microsatellites according to the available technique at each period. Despite the irregularity of genotyping along nearly fifteen years after the new management started, it appears that the proportion of domestic lineage in the river dropped regularly of about 2% each year, a positive result for conservative management measures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The marble trout (Salmo trutta marmoratus) is recognizable at first sight by its very particular marbled skin with no or very few red spots and a high growth potential (Povz 1989; Dorofeyeva et al. 1992; Berrebi et al. 2000; Delling 2002; Simonovic et al. 2007; Sušnik Bajec et al. 2015). It inhabits the north part of Adriatic Sea mainly in the Po, Tagliamento and Isonzo/Soča rivers. It has been subject to intensive researches in evolution and ecology (Berrebi et al. 2000; Crivelli et al. 2000; Snoj et al. 2000; Pustovrh et al. 2011, 2014; Stankovic et al. 2015; Musseau et al. 2015, 2017; Pujolar et al. 2016; Vincenzi et al. 2017). Its mitochondrial sequences are diagnostic (Giuffra et al. 1994; Meraner et al. 2007): based on its mitochondrial control region, this taxon constitutes the MA lineage, one of the 5 main trout lineages which are AT for Atlantic origin, ME for Mediterranean, DA for Danubian, MA for North Adriatic marble trout and AD for Adriatic (Bernatchez et al. 1992). It is also diagnosable with one allozymic marker, the LDH-C1*, with the private allele 120 (with some notable exceptions, see Berrebi et al. 2000).
MA haplotype has been also detected in trout without marbled skin in central Italy (Splendiani et al. 2006), Corsica (Tougard et al. 2018), north and south of the Neretva basin, Croatia and Dalmatia (Bernatchez 2001), Albania (Snoj et al. 2009) and Greece (Apostolidis et al. 1997). Inversely, marbled skin trout inhabits Norway (Skaala et al. 1991; Delling 2002) but this marbled salmonid of the River Otra belongs to the Atlantic brown trout lineage with Atlantic (AT) haplotypes (Skaala and Solberg 1997). It has been shown, by in silico simulations, that the passage from spotted to marbled aspect and vice versa could be a versatile phenomenon, following hybridization for example (Miyazawa et al. 2010). Therefore, both morphological and molecular data are necessary for a clear determination of this taxon. Its taxonomic status is not stabilized, considered as a simple MA lineage inside the brown trout Salmo trutta complex (Bernatchez 2001; Meraner et al. 2007) or as a subspecies S. t. marmoratus (Giuffra et al. 1994, 1996; Lucarda et al. 1999; Nadalin et al. 1999; Pujolar et al. 2011) or a species S. marmoratus (Sommani, 1961; Berrebi et al. 2000; Delling et al. 2000; Snoj et al. 2000; Fumagalli et al. 2002; Pustovrh et al. 2014; Sušnik Bajec et al. 2015). Here, we will consider the marble trout as one of the five main geographic lineages of the S. trutta complex (Bernatchez et al. 1992; Bernatchez 2001) since no convincing molecular phylogeny has still placed the marble trout outside the brown trout.
The marbled trout, with its typical vermiculated aspect, is also the center of interest of anglers from all around Europe and farer (Sullivan et al. 2003) because of its unique coloration pattern and its capacity to grow to more than one meter in particular zones (lakes and reservoirs essentially, from which upstream reproductive migrations have been observed). In Slovenia, it has been established that the main threat for this taxon (and for the linked tourism economy) is its hybridization with lineages of spotted brown trout (Atlantic and Danubian origin) which occurs in northern Adriatic zone (Berrebi et al. 2000; Jug et al. 2005; Meldgaard et al. 2007).
In Slovenia, the main watershed naturally hosting the marble trout is the Soča River, called Isonzo in Italy (Povz 1989). While the Atlantic lineage has an anthropic origin in the Soča watershed, the Danubian lineage have been shown to have a natural ancient origin (Berrebi et al. 2017), which is important to take into consideration for the management of biodiversity protection in the area. Trout management during the last century until 1996 consisted in stocking most parts of the basin with domestic Atlantic domestic strains (Italian then Slovenian strains were used). Molecular data established, in the years 1993–1994, the existence of extensive hybridization resulting, in the main river bed, a more or less equal mixture of allozymic alleles belonging to marble, Danubian and Atlantic brown trout (Berrebi et al. 2000). It has been considered later as a simple hybridization between a natural form, composed of marble lineage naturally crossed with Danubian one, and a domestic lineage, the international Atlantic domestic strain (Berrebi et al. 2017). An upstream–downstream cline of marble-Atlantic lineages has been observed in Soča River, using SNPs (Sušnik Bajec et al. 2015). The upstream end of this cline includes populations of pure marble trout in headwaters of eight tributaries according to allozymes and microsatellites data (Berrebi et al. 2000; Fumagalli et al. 2002). These populations are isolated in small streams separated from the zone of hybridization by impassable waterfalls. In the main Soča River and tributaries, hybridization is still active, described using allozymes, microsatellites and SNPs (Berrebi et al. 2000; Jug et al. 2005; Sušnik Bajec et al. 2015).
The rehabilitation of this iconic taxon has been a stakeholders' target since more than 20 years (Crivelli et al. 2000). The main conservation practices developed by the local Tolmin Angling Association to maintain the marble trout (or at least its phenotype) in the zone of hybridization were: (i) stocking and breeding of the exotic Atlantic brown trout has been banned in all the Soča basin by the Slovenian law in 1996; (ii) stocking with genetically pure or phenotypically selected marble trout strains bred in hatchery; emphasis was placed on stocking of early life stages of marble trout (0 + from 1999 plus eyed eggs from 2007), and (iii) changing regulations for anglers: increasing from 40 cm before 1996 to 50 cm for 1996–2004 and later to 60 cm the limit size of keeping the fished phenotypic marble in the zone of hybridization and maintaining the size limit to 26 cm for brown trout and hybrids.
In the present survey, the evolution of the hybrid proportion is surveyed using allozymes, mitochondrial sequences and microsatellites on a diachronic genetic study over about 20 years (1993–2012) in the middle part of the Soča basin, covering the switch in conservation practices. The main objective is to determine the efficiency of the conservative actions (i.e., banning new stocking of exotic lineages and releasing only phenotypic and pure marble trout produced by local hatcheries). Thus, the quantitative proportions of wild and domestic lineages are measured in order to test if it has been modified after two decades of directional management. Overall, this study allows a prediction of the future of the hybrid population in the Soča basin.
Materials and methods
Sampling
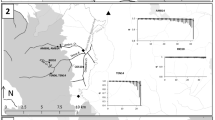
The main sampling effort was focused on a station in the immediate vicinity of the Soča River: the Volarja River, tributary of the former (Fig. 1). This option was decided because sampling the main course of the Soča River is frequently difficult or impossible due to high discharges. Sampling Volarja River is always possible, this is a stable population in a stable river, where restocking has never been practiced and where the trout population is assumed to represent a proxy of the large Soča one with which no obstacle limits the fish movements.
Position of the Volarja station on a simplified map of the five main genetic lineages of brown trout in Europe (according to Bernatchez (2001) and authors' knowledge). In multiples zones, mitochondrial lineages (AT in blue; ME in green, MA in yellow, AD in beige and DA in red) are overlapping each other and produced isolated patches in the range of other lineages: the map shows only the dominant lineages
As detailed in Berrebi et al. (2017), the upper part of the Volarja River is constituted by two converging tributaries called Podbreg and Brinta Rivers, sampled for the present investigation (Table 1). These upstream tributaries of the Volarja River are inhabited by Atlantic domestic trout (Berrebi 2005; Berrebi et al. 2010) introduced by Italian managers at the beginning of the twentieth century. In upstream rivers, a capture-mark-recapture survey (CMR; Lebreton et al. 1992) was performed from 2004 to 2018. Over 2650 trout have been individually tagged in the two upstream tributaries; none have been recaptured downstream (unpublished data), suggesting a very sedentary lifestyle of these trout (Vincenzi et al. 2018). Finally, because the Soča River has been stocked for decades, until 1996, using a commercial domestic Atlantic strain, a sample of a similar strain, from an Isère hatchery (France) has been added as reference (Table 1).
The trout were caught by electro fishing by the Tolmin Angling Association managing the river. For allozymes genotyping, river trout were anaesthetized with MS222 (Western Chemical’s TRICAINE-S) in the field until death, then dissected. The eyes were removed and immediately frozen in liquid nitrogen until analysis. For microsatellites and mtDNA analyses, a small fin clip was removed from phenoxyethanol anaesthetized trout before release in the station of capture. Then, fin tissues are preserved in 96% ethanol for years.
Molecular markers
Along time, several types of markers were used:
Allozymes
The LDH enzymatic system (L-lactate dehydrogenase, E.C. 1.1.1.27), and especially the locus LDH-C1*, has been chosen because it is the only diagnostic marker for marble, Danubian, and Atlantic lineages hybridizing in the Soča River (see Berrebi et al. 2000 for detailed explanations) and because some samples have been genotyped at this unique diagnostic locus. In the laboratory, the tissues samples (eyes) were thawed, homogenized in buffer and centrifuged. The supernatant was used as soluble protein extract. Protein electrophoreses were conducted on horizontal starch gels (12%). LDH enzymatic revealing system and genotyping were established using the staining methods developed by Krieg (1984) and Pasteur et al. (1988), modified in Berrebi et al. (2000).
Mitochondrial DNA (mtDNA)
Using the extracted DNA (see previous and following paragraphs), the mtDNA control region was amplified by PCR using the PST and FST primers of Lee et al. (1995) then characterized by RFLP using the method described by Sušnik Bajec et al. (2015): restriction enzymes allowed distinguishing between MA and DA–AT lineages (AluI) and between AT and DA lineages (SatI).
Microsatellites
DNA was extracted from fin tissue samples using the Chelex/proteinase K protocol described by Walsh et al. (1991) and Estoup et al. (1996). Eight to twelve microsatellite markers were used in this study (BFRO1, BFRO2, Mst15, Mst73, Mst85, Mst543, OmM1105, Omy21DIAS, Oneµ9, Sfo1, Ssa197, SsoSL311), composed of 9 di- and 3 tetra-nucleotide repeat loci, developed in the literature (Table 2) on trout and salmon DNA. For each marker, the 5' end of one of the primers was covalently linked to fluorescein, Cy3 or Cy5 labels. Polymerase chain reactions (PCR) were performed in an Eppendorf Mastercycler programmable thermocycler with a 10 µL reaction volume containing 0.2 U of Taq polymerase (Sigma-Aldrich, Saint-Quentin Fallavier, France), 1.5 to 2 mM MgCl2, 0.4 mM of each dNTP (Invitrogen, St Aubin, France), 1 µL ×10 reaction buffer and 3.75 µM of each primer (Eurofins MWG Operon, Ebersberg, Germany). The thermal cycling conditions were set as follows: initial denaturation (94 °C, 3 min); 30 cycles of denaturation (94 °C, 30 s), annealing (15 s, temperatures given in Table 2) and extension (72 °C, 15 s) cycles; and then a final extension (72 °C, 3 min). The PCR products were electrophoresed in 6% denaturing polyacrylamide gels (Bio-Rad, Marnes-le-Coquette, France) and visualized with a FMBIO-II fluorescent imaging system (Hitachi, Yokohama, Japan). Allele sizes were based on a fluorescently labeled ladder 100–600 bp (Promega, Madison, WI, USA), using the FMBIO Analysis 8.0 image analyzer program (Hitachi, Yokohama, Japan).
Population genetic analyses
Only microsatellite genotypes matrix was analyzed (since allozymes and mtDNA sequences constitute monolocus data each). Because the number of genotyped loci varied each year according to circumstances, three genotypes matrices were constructed and used as a basis for all of the following statistical analyses. The 8-loci matrix included 15 samples and 661 individuals; the 10-loci matrix was composed of 10 samples for a total of 450 individuals; the 12-loci matrix described the genotypes of 374 individuals dispatched in 8 samples. The individuals of the 12-loci matrix are also included in the 10- and the 8-loci matrices. The individuals of the 10-loci matrix belong also to the 8-loci matrix. All the tests were performed three times (for each of the three matrices based on 8, 10 and 12 loci).
The statistical analysis followed a series of tests described below. Before, Micro-Checker 2.2 (Van Oosterhout et al. 2004) was used for genotyping errors detection (null alleles, large allele dropout, scoring of stutter peaks and typographic errors) frequently provoking Hardy–Weinberg deviations .
-
The genetic polymorphism was estimated via expected heterozygosity (He), unbiased estimate of heterozygosity Hnb (Nei 1978), observed heterozygosity (Ho) and mean number of alleles by locus (A) for each sample, using genetix 4.05 (Belkhir et al. 2004). The goal is to know if the evolution of lineages composition of the Soča population is accompanied with polymorphism variation.
-
The intra-sample panmixia (Fis) and the inter-sample differentiation (Fst) were estimated (Weir and Cockerham 2004), using genetix software. In order to provide comparable estimations, the two parameters were only calculated on 8 loci, involving equally all samples. The significance of the Fis and Fst values was evaluated by random permutation procedures (5000 allele permutations within samples for Fis and 5000 individual permutations between samples for Fst). This investigation participates to the estimation of population evolution during two decades. The sequential Bonferroni correction was applied for multiple tests (Rice 1989).
-
A general picture of the trout diversity was described through a principal component analyses (PCA) on the eight microsatellites shared by all sampling groups including Volarja River (sampled in 1993, 2000, 2003 to 2010 inclusively and 2012), Podbreg River (sampled in 2004 and 2008), Brinta River (sampled in 2008) and a fish farm in Isère (sampled in 2008). A total of 661 individuals and 213 alleles were thus analyzed using the software R v3.6.1 with the package adegenet v2.1.3 (Jombart 2008; R core Team 2019). To diminish their impact, missing data (1.1% of the dataset) have been replaced by 0. Ellipses showing 95% confidence interval have been added for each locality in the exception of Volarja River where three ellipses representing the sampling years 1993, 2003 and 2012 have been drawn for further comparison.
-
Bayesian assignment analyses, implemented in STRUCTURE v2.3.3 software (Pritchard et al. 2000), have been applied at the microsatellite multilocus genotypes matrix allowing to estimate the proportion of autochthonous and domestic alleles (Q) along time. The assignment algorithm was run under the admixture model with correlated allele frequencies (Falush et al. 2003). Burn-in period lengths of 100,000 iterations, followed by 200,000 repetitions of the Markov chain Monte Carlo, were chosen. Ten independent runs were performed for each partition tested (K = 1 to 6). The most likely number of genetic clusters in the dataset was estimated with the DeltaK method (Evanno et al. 2005), which was applied using tools available from the STRUTURE HARVESTER website (Earl and von Holdt 2012).
Diachronic analysis
The diachronic survey was based on three categories of markers. Two of them are monolocus: the LDH-C1* allozymic locus and the sequence of the d-loop of the mtDNA. The third one is multilocus, but because of changing circumstances along nearly 20 years of survey, the number of loci varied (8, 10 or 12 loci). The percentages given in the table and in the regression analysis are those obtained with the maximum of loci available for each sample. In order to estimate the evolution of the hybrid population of the Soča basin (through the Volarja River tributary), a multiple linear regression was run using R software v3.6.1 (R core Team 2019). The Atlantic frequencies were estimated along time (Table 1) according to LDH-C1*(90) allele frequencies for allozymes, AT lineage haplotypes frequencies for mtDNA, and domestic assignment frequencies (Q) for microsatellites. Atlantic frequencies were explained by time (i.e., year of capture as a numerical variable) with the type of molecular marker as cofactor.
Each marker was thereafter independently tested using linear regression. Because the microsatellites markers cover a larger number of years sampled, the evolution of genotypes was further investigated through the mean and variance of PC-1 scores obtained from the PCA describe above. Linear regressions have then been performed in the goal to test if means of PC-1 scores could be explained by the sampling year. Since conservation management effort start after 1996, we performed the analysis with and without the sampling year ‘1993’. Standard error of PC-1 scores were computed for each sampling and added on the plot. Then, a polynomial linear regression and a linear regression have been performed to test the relationship between variances of PC-1 scores and sampling year. Polynomial regression has been performed on the complete dataset, while linear regression was performed for the dataset without the sampling year ‘1993’.
Results
The biomolecular genotyping performed each year between 1993 and 2012 fits with the historic scientific progresses (before and after the advent of microsatellites democratization) and to the financial capacities of the team along nearly twenty years. The number of analyzed trout was also depending of the fishing circumstances, the objective being to analyze 30 or more fish. Table 1 summarizes the samples sizes and the molecular analyses performed on each year sample.
Two of the markers analyzed were monolocus and so required no statistical investigation except their evolution along time: allozymes (locus LDH-C1*) genotyped from 1993 to 2003 and D-loop of the mtDNA, from 2003 to 2009. Only microsatellites were analyzed on different aspects as follows. Complete genotype matrices are given as supplementary documents: Table S1 for 8 loci, S2 for 10 and S3 for 12.
We first tested alleles scoring potential errors using Micro-Checker. A total of 156 elementary tests were performed (one test = one locus x one sample). Neither drop-out nor stuttering mistake was detected. On the other hand, possible null alleles were detected 45 times (29% of the tests). Micro-Checker suggested null alleles presence in 38% (44/116) of the tests in Volarja samples, but nearly no (1/32 = 3%) in upper Volarja rivers (Podbreg and Brinta Rivers) and 0% in the Atlantic hatchery sample.
Evolution of genetic diversity
While mostly sampled at the same place, following genetic diversity of a population is precious to detect perturbations. Microsatellite markers were analyzed with population parameters on eleven samples along nineteen years. Table 3 describes, through four parameters (He, Hnb, Ho and A), the diachronic evolution of the diversity. The first calculation reduces the whole data to only 8 loci in order to avoid distortion between 12-, 10- and 8-loci calculations. It appears that the Volarja population suffered a drop of diversity from 0.83 to 0.69 along the survey.
The other diversity parameters were calculated with all the data available. They show similar drop, except the A parameter (mean number of alleles by locus).
The French hatchery, in Isère department, is known to be highly diverse (Bohling et al. 2016). This reference allows considering all value over that of this hatchery as highly polymorphic. Stations upstream the Volarja River are less polymorphic.
Panmixia
Genetic diversity parameters given Table 3 showed a clear discrepancy between the parameters He (or Hnb) and Ho. Globally, these parameters give the mean values of 0.75 and 0.65 respectively, i.e. about 13% of discrepancy, mark of general heterozygote deficiencies. Table 4 confirms this observation with most Fis positive and significant after Bonferroni correction. Only the 2007 Volarja sample, the upstream rivers and the hatchery are in panmixia.
Inter-years differentiation
As observed on genetic diversity, the Volarja River population is fluctuating. The Fst parameter can describe the intensity of the fluctuations. Table 5 shows that rarely two following sampling can be considered as coming from the same population (Fst significantly different from zero). Globally, 5% only of the Fst estimations between Volarja samples can be assimilated to zero, never between following years. Most of the comparisons are highly significantly different to zero, as if they were sampled on different populations.
Multidimensional picture
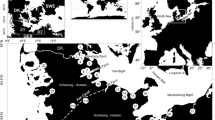
The PCA shows a clear distinction between each locality (Fig. 2) except between the two upstream populations. Interestingly, PC-1 axis reveals that Volarja River individuals sampled in older campaigns show a tendency to be closer to the upstream populations on PC-1 axis. PC-2 principally shows genetic differentiation between fish farm of Isère and wild populations. Thus, only PC-1 scores will be used for further analyses (see below). Finally, 95% confidence interval support a clear genetic difference between each population in addition to diminution in genetic variance in more recent sampling in the Volarja River (Fig. 2-right) parallel to the drop of diversity (Table 3).
Left: PCA describing the genetic diversity contained in all samples genotyped at 8 microsatellites loci (213 alleles) for 661 individuals from 4 localities. Ellipse represents 95% confidence interval. Right-top: Relationship between mean of PC-1 scores and time with standard error bars for Volarja samples only. Right-bottom: Relationship between variance of PC-1 scores and time in Volarja station. Dotted line in relationship has been provided on all dataset while solid line is the relationship for years after 1996, where conservative management effort starts
Assignment analyses
Three series of test were performed analyzing the three matrices.
The best fitting suggested by the DeltaK method was 2 for the three tests, meaning that two main clusters (natural and domestic) of populations were identified. For K = 3, ancient Atlantic strains (upstream) and present commercial one (French hatchery) are differentiated. The diagram (not shown) indicates that the ancient strain is not present in the Volarja population. The three consensus values of assignment (based on the mean Q values of the ten tests performed for each matrix) are shown in Table 6 and Fig. 3. There is very limited difference for a given sample when analyzed with 8, 10 or 12 loci. The assignment analysis of sample 9 (Volarja River in 2005) showed the larger departure: the Atlantic domestic proportion changed from 0.25 with 8 loci to 0.29 for 10 and 0.27 for 12 loci (Table 6). This difference reaching 15% is however not really visible in Fig. 3.
Regression analyses
Table 1 gives the proportion of domestic trout (i.e. of Atlantic origin) in the Volarja River, in two of its upstream tributaries and of the current commercial domestic trout strain of the Atlantic lineage. The detailed assignment results given Table 6 show the different estimations according to the number of loci.
The multiple linear regression show, after controlling by marker types as cofactor, a significant effect of sampling years on the frequency of AT (adjusted R2 = 0.24, estimate = -0.013, F = 9.62, P = 0.01; Fig. 4; black line). When testing each marker separately with linear regression, mtDNA and allozyme datasets do not show significant results (P = 0.981 and 0.117 respectively), while microsatellites were significant and show that AT frequency variation was mainly explained by the variable time (adj. R2 = 0.95, estimate = -0.021, F = 160.67, P < 0.001; Fig. 4; red dotted line). Note here that, in order to know if the 1993 values (with especially high proportion of AT alleles) are alone responsible of the significance of the reduction of domestic alleles, the association remains similar after removing data from the year 1993 (adj. R2 = 0.82, estimate = -0.020; F = 37.48, P < 0.001; Fig. 4). Together, this suggests that the Atlantic domestic frequency drop of 1.3% (all markers), to 2.1% (only microsatellites) per year.
Plot of Atlantic lineage frequencies (y axis) presented against time (x axis). In black dots, the Atlantic presence is estimated by the LDH-C1 allozyme; in green by mtDNA sequences and in red by 8 to 12 microsatellite loci. The black line is calculated from all the markers and the red dotted line from microsatellites only
Further investigation on microsatellites genotypes surrogated by PC-1 scores shows a clear evolution of genotype in Volarja River (adj. R2 = 0.70, F = 23.93, P < 0.001; Fig. 2-right), differentiating from other wild populations with Atlantic origins (Fig. 2-left). This relationship shows an increase in genetic variation explained, through the adjusted R2 statistic, when removing the sampling year ‘1993’ (adj. R2 = 0.82, F = 41.86, P < 0.001; Fig. 2-right). Finally, a significant decrease in variance is also observed over time when excluding the sampling year ‘1993’. (adj. R2 = 0.35, F = 5.80, P = 0.043; Fig. 2-right).
Discussion
Today, seven genetically pure marble trout populations are located in headwaters (Fumagalli et al. 2002), after the loss of the eighth following a catastrophic landslide; in total 14.7 km of stream are inhabited with genetically pure marble trout in the Adriatic basin of Slovenia (Crivelli and Jesenšek, unpublished data). All the remaining streams and rivers of the upper Soča basin, the zone of hybridization, shelter only hybrid trout. The conservation objective was to restore the marble genes in this zone.
The rehabilitation of the marble trout has been a stakeholders' target since more than 20 years (Crivelli et al. 2000), following the Allendorf et al. (2001) recommendations which are (i) not stocking with exotic strains but with genetically pure or phenotypically selected marble trout strains in hatchery and (ii) change regulations for anglers. Adapting management practices in order to save the marble trout is a valuable action helping biodiversity conservation, but verifying the impacts of the methods used is essential to confirm their effectiveness (Schwartz et al. 2006). That is why, since 1993, regular captures and analyses of trout of the Soča catchment were performed until 2012. Sharing the results with the scientific community will thus be of interest for any conservation management group attempting, after decades of stocking, to return to an autochthonous fish lineage.
The difficulty to overcome on this long-term survey was the disparity of the markers used (depending of the scientific history of molecular progresses, Allendorf 2017) and the financial limitations encountered along this two decades period. Table 1 gives a clear picture of what have been done in terms of markers and samples sizes. Allozymes were first used, then mtDNA and microsatellites. For the two last markers, it has been possible to re-use preserved tissues of the 1993, 2000 and 2003 samples.
Survey strategy
Diachronic surveys cannot be organized like classical analysis of a population or of the geographic organization of a taxon. Long term surveys must maintain the effort during years (here 20 years), with field campaigns and laboratory work, with grants corresponding to both field and laboratory expenses. Along the two decades, the first objective was to understand the genetic structure of the Soča River populations. The last input of the 2017 survey (Berrebi et al. 2017) are (i) the confirmation that the Volarja River station is representative of the main Soča one, (ii) that the marble trout naturally hybridized with Danubian ones long ago at a rate probably harmless to the particular marbled external morphology, and finally (iii) that Atlantic domestic lineage was recently stocked and caused the appearance of hybrid phenotypes gathering vermiculation and spots forming diverse colored aspects. Microsatellites cannot discriminate marble and Danubian origin (Fig. 2 and Fig. 3 in Berrebi et al. 2017). Probably because of its high mutation rate, only recent phenomena can be detected by this hypervariable marker. Here, only natural/domestic lineages are so observed. Consequently, following Atlantic lineage with the three markers (Fig. 4) is the simplest method to record a genetic evolution during the conservative management.
Hardy–Weinberg disequilibria
According to Table 4, a general deficit of heterozygote genotypes covers all Volarja samples (except one in 2007), but no deficit is detected in upstream samples (Podbreg and Brinta Rivers). Similarly, according to Micro-Checker output, numerous null alleles are suspected in Volarja samples, affecting different loci each year, but this is not observed in upstream stations (except one) nor in the Atlantic hatchery strain.
The present pattern rather suggests a Wahlund effect caused by continuous stocking: only Volarja samples show heterozygotes deficit, not in its two upstream tributaries (Table 4: Podbreg and Brinta Rivers). Null alleles should not disappear in the Volarja population for a given locus depending on the years of sampling.
Before 1996, stocking was practiced with domestic Atlantic and marble strains and wild phenotypic marble trout (i.e. pure marble phenotypes were used for fry production then stocked in the Soča, with probable Danubian and Atlantic alleles in low frequencies). More recently, while Atlantic strains were banned, domestic marble trout produced from upstream isolated pure marble populations and phenotypic marble trout descents were continuously stocked. This continuous introduction of various stocks probably provoked the detected disequilibria; this is not observed in upstream tributaries, not stocked since a century, isolated by impassable obstacles.
A decrease of Atlantic lineage
According to the geographic structuration of the S. trutta complex (Bernatchez 2001), the Atlantic lineage detected in the Soča River is exotic and so due to ancient or recent introductions. Until the 1996 banning, hatcheries rearing the Atlantic domestic trout were active in Slovenia, producing fry for stocking. Ancient introduction in Soča tributaries at the time the region was Italian have been documented. The first recorded introductions in Soča River date back to 1906 (Povz et al. 1996). Later, the management of the rivers included extensive stocking with brown trout resulting in hybridization (Jesenšek 1994; Povz et al. 1996). The present study clearly established that since 1996, the Atlantic lineage decreased continually, the microsatellites estimation mostly following the regression line (Fig. 4). If the three kinds of markers are used, the decrease is of about 1% the year. With microsatellites only, the Atlantic proportion decreased from 42 to 13%, inducing a significant estimation of about 2% decrease per year. In addition to this genetic evolution, the diversity in Soča River population shows a diminution with time (Table 3). Considering that hybridization generally increase genetic diversity inside a population, it is thus expected that a decrease in variation will be associated to a purge of exogenous genetic variants.
Fst analysis provides similar information. Table 5 is divided into two sorts of values. While generally significant, the Fst estimation, based on 8 loci, between pairs of Volarja samples are low to very low (0.00 < Fst < 0.08) but values between Volarja samples and Atlantic ones (i.e. both upstream stations and the French hatchery) are high to very high (0.13 < Fst < 0.33). On the other hand, along with the diachronic reduction of the Atlantic alleles, the Fst estimates, based on 8 loci, between the Volarja and the Atlantic samples grow steadily as expected (Fig. 5).
In another salmonid (brook charr: Salvelinus fontinalis), time since the end of restocking was also a main factor explaining the level of hybridization, suggesting that genetic drift and selection can both act together with the possibility to a return to a nearby wild genetic state after stocking cessation (Létourneau et al. 2018; Leitwein et al. 2020). In addition, presence of deleterious recessive mutations was strongly associated to the recombination rates in stocked wild populations (Leitwein et al. 2020). Based on these results, the authors suggest that stocking had the potential to reduce deleterious mutation frequencies in wild population. In parallel, Ferchaud et al. (2018) observed that smaller populations show higher deleterious mutation frequencies in unstocked wild Lake Trout (S. namaycush), but such relationship was not present in stocked populations. However, all stocked populations of Lake Trout in the same area loss their genetic integrity and cluster together in assignment analyses according to the source of stocking (Valiquette et al. 2014). Altogether, this suggests that stocking can help smaller populations with conservation issue to purge deleterious mutations but the source of stocking should also be locally adapted in order to preserve the genetic integrity of the stocked population.
The given examples support the idea that Atlantic lineage continuously decreases over time in the Soča River. The extrapolation of the slope (Fig. 4) leads to a disappearance of Atlantic alleles around 2020, which is a very optimistic prospective. A scenario in which the Atlantic lineage remains at a low level is more realistic. Of course, next step will be to verify this almost disappearance expectation, but in any cases, the decrease in AT lineage can only be due to the conservation management practices applied for decades in the Soča basin. Genetic control of threatened taxon management is expensive; however field conservation success can be obtained without major funding sources. With good conservation practices knowledges, any small conservation management group around the world can protect their fish biodiversity.
Availability of data and material
The data are available on simple request to the corresponding author.
Code availability
Not applicable.
References
Allendorf FW (2017) Genetics and the conservation of natural populations: allozymes to genome. Mol Ecol 26:420–430
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:613–622
Angers B, Bernatchez L, Angers A, Desgroseillers L (1995) Specific microsatellite loci for brook charr reveal strong population subdivision on a microgeographic scale. J Fish Biol 47(Supplement A):177–185
Apostolidis AP, Triantaphyllidis C, Kouvatsi A, Economidis PS (1997) Mitochondrial DNA sequence variation and phylogeography among Salmo trutta L. (Greek brown trout) populations. Mol Ecol 6:531–542
Belkhir K, Borsa P, Goudet J, Bonhomme F (2004) GENETIX 4.05: Logiciel sous Windows pour la génétique des populations, Laboratoire Génome et Population, CNRS-UPR, Université de Montpellier II: Montpellier, France, 2004
Bernatchez L (2001) The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 55:351–379
Bernatchez L, Guyomard R, Bonhomme F (1992) DNA sequence variation of the mitochondrial control region among geographically and morphologically remote European brown trout Salmo trutta populations. Mol Ecol 1:161–173
Berrebi P (2005) Suivi de la composition génétique des truites de la Volarja (Slovénie) entre 1993 et 2004—Analyses par allozymes et microsatellites. Research report, Université Montpellier 2. https://data.oreme.org/trout/home
Berrebi P, Povz M, Jesenšek D, Crivelli AJ (2000) The genetic diversity of native, stocked and hybrid populations of marble trout in the Soča river, Slovenia. Heredity 85:277–287
Berrebi P, Cattaneo-Berrebi G, Lasserre B, Dubois S, Shao Z, Zenboudji S, Tougard C (2010) Rapport Volaja 2010: où en sommes-nous après 17 ans d'analyses génétiques? Research report, Université Montpellier 2. https://data.oreme.org/trout/home
Berrebi P, Jesenšek D, Crivelli AJ (2017) Natural and domestic introgressions in the marble trout population of Soča River (Slovenia). Hydrobiologia 785:277–291
Bohling J, Haffray P, Berrebi P (2016) Genetic diversity and population structure of domestic brown trout (Salmo trutta) in France. Aquaculture 462:1–9
Crivelli AJ, Poizat G, Berrebi P, Jesenšek D, Rubin J-F (2000) Conservation biology applied to fish: the example of a project for rehabilitating the marble trout (Salmo marmoratus) in Slovenia. Cybium 24:211–230
Delling B (2002) Morphological distinction of marble trout, Salmo marmoratus, in comparison to marbled Salmo trutta from River Otra, Norway. Copeia 26:283–300
Delling B, Crivelli A, Rubin J-F, Berrebi P (2000) Morphological variation in hybrids between Salmo marmoratus and alien Salmo species in the Volarja stream, Soča River basin, Slovenia. J Fish Biol 57:1199–1212
Dorofeyeva YA, Vukovich T, Seratlich D (1992) Osteological features of the endemic Balkan Marbled trout, Salmo marmoratus Cuv. (Salmonidae). Ichthyologia 31:113–121
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Estoup A, Largiader CR, Perrot E, Chourrout D (1996) Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Mol Mar Biol Biotechnol 5:295–298
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Ferchaud AL, Laporte M, Perrier C, Bernatchez L (2018) Impact of supplementation on deleterious mutations distribution in an exploited salmonid. Evol Appl 11:1053–1065
Fumagalli L, Snoj A, Jesenšek D, Balloux F, Jug T, Duron O, Brossier F, Crivelli AJ, Berrebi P (2002) Extreme genetic differentiation among the remnant populations of marble trout (Salmo marmoratus) in Slovenia. Mol Ecol 11:2711–2716
Giuffra E, Bernatchez L, Guyomard R (1994) Mitochondrial control region and protein coding genes sequence variation among phenotypic forms of brown trout Salmo trutta from northern Italy. Mol Ecol 3:161–171
Giuffra E, Guyomard R, Forneris G (1996) Phylogenetic relationships and introgression patterns between incipient parapatric species of Italian brown trout (Salmo trutta L. complex). Mol Ecol 5:207–220
Holm LE, Bendixen C (2000) Oncorhynchus mykiss clone TAA72-13. Sequence tagged site. Accession number AF239038
Jesenšek D (1994) Artificial propagation of marble trout (Salmo marmoratus Cuvier 1817) in the Fisheries Society of Tolmin. In: Kirchhofer A, Hefti D (eds) Symposium on the conservation of endangered freshwater fish in Europe. Bern, Switzerland: University of Bern, SSHL, FOEFL, 24
Jombart T (2008) adegene"t: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jug T, Berrebi P, Snoj A (2005) Distribution of non-native trout in Slovenia and their introgression with native trout populations as observed through microsatellite DNA analysis. Biol Conserv 123:381–388
Krieg F (1984) Recherche d'une différenciation génétique entre populations de Salmo trutta: Université de Paris-Sud, Orsay
Lebreton J-D, Burnham KP, Clobert J, Anderson DR (1992) Modelling survival and testing biological hypothesis using marked animals: a unified approach with case studies. Ecol Monogr 62:67–118
Lee HS, Conroy J, Howell WH, Kocher TD (1995) Structure and evolution of teleost mitochondrial Control Regions. J Mol Evol 41:54–66
Leitwein M, Duranton M, Rougemont Q, Gagnaire P-A, Bernatchez L (2020) Using haplotype information for conservation genomics. Trends Ecol Evol 21:4755–4769
Létourneau J, Ferchaud A-L, Le Luyer J, Laporte M, Garant D, Bernatchez L (2018) Predicting the genetic impact of stocking in Brook Charr (Salvelinus fontinalis) by combining RAD sequencing and modeling of explanatory variables. Evol Appl 11:577–592
Lucarda NA, Bargelloni L, Patarnello T (1999) Genetic characterisation of Salmo trutta marmoratus (Cuvier, 1817) populations by means of nuclear markers: preliminary results. Quad Ente Tutela Pesca - Udine 28:1–5
Meldgaard M, Crivelli AJ, Jesenšek D, Poizat G, Rubin J-F, Berrebi P (2007) Hybridization between the endangered marble trout (Salmo marmoratus) and the brown trout (Salmo trutta): an in-stream experiment. Biol Conserv 136:602–611
Meraner A, Baric S, Pelster B, Dalla Via J (2007) Trout (Salmo trutta) mitochondrial DNA polymorphism in the centre of the marble trout distribution area. Hydrobiologia 579:337–349
Miyazawa S, Okamoto M, Kondo S (2010) Blending of animal colour patterns by hybridization. Nat Commun 1:66
Musseau C, Vincenzi S, Jesenšek D, Cantera I, Bouletreau S, Santoul F, Crivelli AJ (2015) Direct and indirect effects of environmental factors on dietary niches in size-structured populations of a wild salmonid. Ecosphere 6(12):256
Musseau C, Vincenzi S, Jesenšek D, Crivelli AJ (2017) Cannibalism in a non-native brown trout Salmo trutta and rainbow trout Oncorhynchus mykiss stream-dwelling populations. J Fish Biol 91:1737–1744
Nadalin G, Battistella S, Amirante GA (1999) Preliminary considerations on nuclear DNA as a genetic marker for marble Trout (Salmo trutta marmoratus). QuadEnte Tutela Pesca - Udine 28:13–16
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
O'Reilly PT, Hamilton LC, McConnell SK, Wright JM (1996) Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotids and tetranucleotids microsatellites. Can J Fish Aquat Sci 53:2292–2298
Pasteur N, Pasteur G, Bonhomme F, Catalan J, Britton-Davidian J (1988) Practical isoenzyme genetics. Halstred Press, New York
Povz M (1989) Distribution and biometric characteristics of the marble trout (Salmo marmoratus Cuvier 1817) in Slovenia. Ichthyos 8:29–36
Povz M, Jesenšek D, Berrebi P, Crivelli AJ (1996) The Marble trout, Salmo trutta marmoratus, Cuvier 1817 in the Soča River basin, Slovenia. Tour du Valat Publication, Le Sambuc
Presa P, Guyomard R (1996) Conservation of microsatellites in three species of salmonids. J Fish Biol 49:1326–1329
Presa P, Krieg F, Estoup A, Guyomard R (1994) Diversité et gestion génétique de la truite commune: apport de l'étude du polymorphisme des locus protéiques et microsatellites. Genet Sel Evol 26(suppl.1):183–202
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pujolar JM, Lucarda AN, Simonato M, Patarnello T (2011) Restricted gene flow at the micro- and macro-geographical scale in marble trout based on mtDNA and microsatellite polymorphism. Front Zool 8:7
Pujolar JM, Vincenzi S, Zane L, Crivelli AJ (2016) Temporal changes in allele frequencies in small marble trout Salmo marmoratus population threatened by extreme flood events. J Fish Biol 88:1175–1190
Pustovrh G, Sušnik Bajec S, Snoj A (2011) Evolutionary relationship between marble trout of the northern and the southern Adriatic basin. Mol Phylogenet Evol 59:761–766
Pustovrh G, Snoj A, Sušnik Bajec S (2014) Molecular phylogeny of Salmo of the western Balkans, based upon multiple nuclear loci. Genet Sel Evol 46:7
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/
Rexroad CE, Coleman RL, Hershberger WK, Killefer J (2002) Rapid communication: thirty-eight polymorphic microsatellite markers for mapping in rainbow trout. J Anim Sci 80:541–542
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Schwartz MK, Luikart G, Waples RS (2006) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22(1):25–33
Scribner KT, Gust JR, Fields RL (1996) Isolation and characterization of novel salmon microsatellite loci: cross-species amplification and population genetic applications. Can J Fish Aquat Sci 53:833–841
Simonovic P, Maric S, Nikoloc V (2007) Trout Salmo spp. complex in Serbia and adjacent regions of the western Balkans: reconstruction of evolutionary history from external morphology. J Fish Biol 70(Suppl. C):359–380
Skaala O, Solberg G (1997) Biochimical genetic variability and taxonomy of a marmorated salmonid in River Otra, Norway. Nordic J Freshw Res 73:3–12
Skaala O, Jorstad KE, Borgstrom R (1991) Fine-spotted brown trout: genetic aspects and the need for conservation. J Fish Biol 39(A):123–130
Slettan A, Olsaker I, Lie Ø (1995) Atlantic salmon Salmo salar microsatellites at the SSOSL25. SSOSL85. SSOSL311. SSOSL417 loci. Anim Genet 26:277–285
Snoj A, Pohar J, Dovc P (1997) The first microsatellite DNA marker for marble trout, BFRO 001. J Anim Sci 75:1983
Snoj A, Jug T, Melkic E, Sušnik S, Pohar J, Dovc P (2000) Mitochondrial and microsatellite DNA analysis of marble trout in Slovenia. Quad Ente Tutela Pesca - Udine 29:5–11
Snoj A, Maric S, Berrebi P, Crivelli AJ, Shumka S, Sušnik S (2009) Genetic architecture of trout from Albania as revealed by mtDNA control region variation. Genet Sel Evol 41:22
Sommani E (1961) Il Salmo marmoratus Cuv. : sua origine e distribuzione nell’Italia settentrionale. Bolletino Pesca Piscic Idrobiol 15:40–47
Splendiani A, Giovannotti M, Cerioni PN, Caniglia ML, Caputo V (2006) Phylogeographic inferences on the native brown trout mtDNA variation in central Italy. Ital J Zool 73:179–189
Stankovic D, Crivelli AJ, Snoj A (2015) Rainbow trout in Europe: introduction, naturalization, and impacts. Rev Fish Sci Aquac 23:39–71
Sullivan CA, Jesenšek B, Jesenšek D, Zuza A (2003) An assessment of the importance of recreational sports fishing in the upper Soča basin, Slovenia. Project report to Tour du Valat, CEH, Wallingford, UK
Sušnik S, Snoj A, Pohar J, Dovc P (1997) The microsatellite marker (BFRO 002) characteristic for different geographically remote brown trout, Salmo trutta L., populations. Anim Genet 28:370–383
Sušnik Bajec S, Pustovrh G, Jesenšek D, Snoj A (2015) Population genetic SNP analysis of marble and brown trout in a hybridization zone of the Adriatic watershed in Slovenia. Biol Cons 184:239–250
Tougard C, Justy F, Guinand B, Douzery EJP, Berrebi P (2018) Salmo macrostigma (Teleostei, Salmonidae): nothing more than a brown trout (S. trutta) lineage? J Fish Biol 93:302–310
Valiquette E, Perrier C, Thibault I, Bernatchez L (2014) Loss of genetic integrity in wild lake trout populations following stocking: Insights from an exhaustive study of 72 lakes from Québec, Canada. Evol Appl 7:625–644
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER : software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vincenzi S, Mangel M, Jesenšek D, Garza JC, Crivelli AJ (2017) Genetic and life-history consequences of extreme climate events. Proc R Soc Lond B Biol 284:2016–2118
Vincenzi S, Jesenšek D, Crivelli AJ (2018) A framework for estimating the determinants of spatial and temporal variation in vital rates and inferring the occurrence of unobserved extreme events. R Soc Open Sci 5:171087
Walsh PS, Metzger DA, Higushi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgements
The authors warmly thank Ghislaine Cattaneo-Berrebi, Bernard Lasserre, Sophie Dubois and Zhaojun Shao for genotyping most of the trout in the genetic laboratory of the Montpellier University, and David Schikorski of the Labofarm private company (France) for genotyping another part of the trout. This research was partly funded by MAVA Foundation to A.J.C.
Funding
This research was funded by MAVA Foundation to A.J.C.
Author information
Authors and Affiliations
Contributions
All authors contributed equally.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Ethics approval
Sampling was done at a time no administrative authorization was necessary. However, because all the captures were done by local managers (Tolmin Anglers Association), it was done in agreement with local authorities. Concerning fish sacrifice, samples for allozymic genotyping were anesthetized with MS222 until death. This is one of the authorized methods.
Consent to participate
All four authors consent to participate.
Consent for publication
All four authors consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Berrebi, P., Jesenšek, D., Laporte, M. et al. Restoring marble trout genes in the Soča River (Slovenia). Conserv Genet 23, 481–494 (2022). https://doi.org/10.1007/s10592-022-01430-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-022-01430-0