Abstract
Land conversion is one of the major global changes that threaten population viability. As with many industrial activities, quarrying highly modifies land cover, destroying previous habitats but also creating new conditions potentially supporting functioning and connectivity of pioneer species. Using a multi-landscape and -temporal approach, we assessed the impact of quarrying on the genetic diversity of two amphibians with contrasted ecological constraints: the common toad (Bufo bufo) and the natterjack toad (Bufo calamita), favouring vegetated and pioneer environments, respectively. The study was conducted across six areas of ca. 250 km2 each. Mixed effect models were used to determine which landscape features affect the genetic diversity of the two species. These analyses were performed at three time points (1940s, 1970s and 2000s). Genetic diversity of B. bufo was found to increase with the area of semi-wooded and herbaceous vegetation, and decrease with the area of roads and urbanized areas. Genetic diversity of B. calamita increased with the area of bare ground and of quarries, and decreased with the area of dense woods. We found no effect of quarrying on B. bufo, unlike for B. calamita in which genetic diversity was favoured by quarrying at all three time-points. Despite having similar generation times, B. bufo’s diversity was best explained by 1940s landscape and that of B. calamita by 2000s landscape. This study enlightens the genetic conservation value of quarries for pioneer species and the possible delays between landscape changes and their effects on the populations of some, but not all, species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic diversity is one of the fundamental levels of biodiversity. It influences a number of ecological processes (Hughes et al. 2008) and provides the raw material that allows species to adapt to changing environmental conditions (Frankham et al. 2010). Genetic diversity is therefore especially important in the context of adaptation to global changes. Besides mutational processes, genetic diversity depends mainly on population size and gene flow (Leimu et al. 2006): decrease in population size results in lower genetic diversity through increased genetic drift, whereas incoming gene flow is a source of genetic variability. Genetic diversity thus encapsulates both population demography and landscape connectivity, the main processes by which anthropogenic land conversion may affect population functioning and viability.
Anthropogenic land cover conversion is currently acknowledged as one of the major drivers of genetic erosion worldwide. Through habitat loss and fragmentation, land conversion reduces overall habitat availability and favours the isolation of suitable patches (Wilcox and Murphy 1985; Hanski 1999; Fahrig 2003). This leads to decreased environment quality and loss of landscape connectivity (i.e., the degree to which a landscape facilitates or impedes movements among resource patches; Taylor et al. 1993). In Europe, intensive anthropogenic land conversion is mainly driven by urbanization, development of transportation infrastructures, intensive farming and logging (Martino and Fritz 2008). While the impact on genetic diversity of many of those activities has been extendedly studied (e.g., urbanization: Blanchong et al. 2013; infrastructure transportation: Epps et al. 2005; forestry: Dixo et al. 2009) others remain unknown, as is the case for quarrying activities.
Quarrying, the industrial extraction of mineral materials used in construction, impacts small areas at the regional scale (e.g., less than 1 % of French national territory, Barnaud and Le Bloch 1998), but high activity concentration can result in severe modifications of land cover at the local scale (e.g., valleys). Extracting activity results in the replacement of the original ecosystems by bare soil, with increased dust, noise and traffic, and may by this way degrade the local environment (Clements et al. 2006; Berhe 2007; Lameed and Ayodele 2010). Resulting topsoil is generally thin, nutrient-poor and compacted: successional processes cannot proceed and are stuck at early stages of colonization. Interestingly, though, these profound alterations result in the creation of xeric and pioneer habitats. Artificial walls, bottom, barren terraces, installation plants and stocks have been observed to attract specialist species in every taxa: invertebrates (Brandle et al. 2000; Benes et al. 2003), amphibians (Rannap et al. 2007), birds (Kovacs 2001; Voetzel et al. 2008) and flora (Novak and Konvicka 2006). In addition, operating gravel pits often generate mosaics of waterbodies that spread across dozens of kilometres and may act as refuge for threatened freshwater fauna and flora (Santoul 2000; Santoul et al. 2009). Also, after exploitation, portions of quarrying sites are rehabilitated (for environmental, visual and safety reasons) into a large panel of ecosystems including heathlands, wetlands or forests (Schulz and Wiegleb 2000; Bzdon 2008; Tropek and Konvicka 2008). This diversity of specific ecosystems gives quarries a high potential for biodiversity conservation. Nonetheless, the effects of quarries on genetic diversity have not been investigated yet and remain unclear: on one hand quarrying could be detrimental to species sensitive to industrial impact by increasing the fragmentation of their habitat, while, on the other hand, it could benefit pioneer species through the creation of new habitat and enhanced connectivity. In this study, we assessed the effects of quarries on genetic diversity relative to the effects of other landscape features in two amphibian species with contrasting ecology.
Whereas land conversion potentially threatens all taxa, amphibians undoubtedly constitute one of the most impacted ones (Cushman 2006; Hof et al. 2011). This sensitivity is mainly due to their poor dispersal capacity (Sinsch 1990; Smith and Green 2005), specific habitat requirements (Karraker and Gibbs 2009), and complex life cycle involving spatially distinct breeding and foraging habitats (Pope et al. 2000). Our two study species are the natterjack toad (Bufo calamita), a specialist of pioneer habitats, and the common toad (Bufo bufo), which favours densely vegetated environments. To provide a complete diagnostic of the effect of particular landscape elements on population functioning, it is highly recommended to use comparative approaches involving species with distinct biological requirements, multiple landscapes and taking land use history into account (Holzhauer et al. 2006; Zellmer and Knowles 2009; Goldberg and Waits 2010). We hence conducted our study at six distinct sites in France, characterized by contrasted landscapes in terms of land use and quarrying activity. We also used several temporal representations of the landscape (i.e., mid 1940s, 1970s and 2000s), to account for potential delays in the effects of landscape changes and their effects on genetic diversity. We predicted quarries to have a negative impact on the genetic diversity of the species favouring forests (B. bufo), and a positive effect on the pioneer natterjack toad (B. calamita).
Materials and methods
Study species
The common toad (B. bufo) is a widespread anuran in Europe. It can be observed in a large panel of terrestrial habitats such as hedgerows, pastures or suburban gardens, but shows preference for forests, both for home-ranging and for dispersal (Denton 1991; Denton and Beebee 1994; Hartel et al. 2008; Janin et al. 2009). High mortality has been observed in open and sandy areas such as those encountered in active quarries (Denton and Beebee 1991, 1994), illustrating its vulnerability to arid and poorly-vegetated habitats (Romero and Real 1996). Breeding is more conditioned by landscape quality than pond quality and configuration (Scribner et al. 2001; Hartel et al. 2008). If dispersal distances have been recorded up to 3 km (Smith and Green 2005), migration distances usually fall within 50–1,600 m (Glandt 1986; Sinsch 1989). Adults exhibit high site fidelity (79–96 % of adults return to the pond they used the previous year; Reading et al. 1991), suggesting that dispersal events are mainly due to juveniles (up to 20 % of the toadlets disperse away from their native pond, Schlupp and Podloucky 1994; Cooke and Oldham 1995). Generation time is around 3 years (Halley et al. 1996).
The natterjack toad (B. calamita) is an endemic species of central and western Europe that shows strong specialisation to poorly-vegetated habitats such as coastal meadows, dunes or heathlands. Breeding occurs in shallow ephemeral ponds with low nutrient levels (Denton 1991; Beebee 1983; Romero and Real 1996). Sandy soils are preferred because of the possibility to dig burrows, which decreases the risk of desiccation (Denton and Beebee 1994). This species also exhibits high fidelity to breeding areas (Sinsch 1992, 1997; Denton and Beebee 1993), and most dispersal events seem to be performed by juveniles (Sinsch 1997). Higher movement speed and distance by toadlets have been observed on open habitat (Stevens et al. 2004), where perception and hunting are more efficient than in densely-vegetated conditions (Denton and Beebee 1994). However, forests seem to be preferentially used by toadlets for dispersal (Stevens et al. 2006). The range of dispersal distances is larger than that of B. bufo, as maximum distances of up to 4 km were most often recorded (Smith and Green 2005) and occasionally up to 12 km (Sinsch et al. 2012). Generation time is thought to be around 3 years (Rowe et al. 2000). The overall decline of natterjack populations across Europe is largely due to the dramatic loss of breeding habitat, which forces this species to colonize substitution environments such as quarries, construction sites or port areas (Eggert and Miaud 2004; Stevens et al. 2006; Rannap et al. 2007).
Study sites
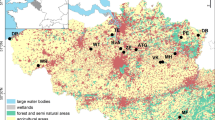
The study was carried out at 6 sites located in northern France and that showed contrasting landscapes and quarry densities (Fig. 1 and supplementary material S1). All sites are rural landscapes with relatively low human densities. They also exhibit similar overall flat topography, with low average elevations (ca. 10–140 m). The northernmost sites-Anneville (A), Poses-Bouafles (P) and Guernes (G) are spread across the Seine river valley, where balanced proportions of forests and farmland characterize the landscape. The Seine has a channelized riverbed. The site of Larchant (L) is located in the southern limit of the wide forested continuum of Fontainebleau and the beginning of the open-field plateau of the Beauce. In the very center of the Beauce, Voves (V) shows a homogeneous open-field landscape. The southernmost site Bonnee (B) is located in the Loire valley, characterised by balanced proportions of forests, farmlands but also semi natural heathlands on the banks (a result of non-channelized riverbeds).
Sampling
We sampled fertilized eggs and larvae during the springs of 2011 and 2012. To minimize sib-effect, we collected eggs from distinct clutches, and when possible larvae in separate ponds: females of both species generally lay one clutch every year (Denton and Beebee 1996; Hitchings and Beebee 1998) that is fertilized by one single male (Banks et al. 1994; Hitchings and Beebee 1998). Once collected, we reared clutch fragments in the laboratory at 20 °C until tadpoles were larger than 1 cm. Individuals were euthanized and stored in 70 % ethanol at room temperature.
Genetic data collection
We extracted DNA from tissue samples using the NucleoSpin® 96 Tissue Kit (Macherey–Nagel) following an automated processing on a robotic platform epMotion 5075 (Eppendorf). We tested fifteen species-specific microsatellite markers for B. bufo (Brede et al. 2001) and eighteen for B. calamita (Rowe et al. 1997, 2001; Rogell et al. 2005). Five PCR procedures were required (see details in supplementary material S2). After extensive tests with different PCR protocols, we retained eleven markers for B. bufo and thirteen for B. calamita (supplementary materials S2). The samples were genotyped using an ABI PRISM 3130 automated sequencer (Applied Biosystems). We screened allele sizes relative to standard marker LIZ 500 (Applied Biosystems) using GeneMapper v4.0 (Applied Biosystems).
Genetic data quality
We controlled genetic data quality following Carlsson et al. (2004). Each run of 96 PCRs included eight positive controls with known genotypes to control for potential contamination and electrophoresis conditions. We checked genotypes for high null allele frequencies, stuttering and large allelic dropout with MICRO-CHECKER v2.23 (Van Oosterhout et al. 2004). We tested linkage disequilibrium between all pairs of loci using GENEPOP v4.2 (Raymond and Rousset 1995; Rousset 2008). Markov chain parameters included a dememorization step of 10,000 iterations followed by 1,000 batches of 10,000 iterations each. We evaluated deviations from Hardy–Weinberg equilibrium using GENEPOP v4.2 (Rousset 2008). Significance levels were calculated with the Markov chain method, with 10,000 dememorization steps, and 100 batches with 5,000 iterations per batch. We accounted for multiple comparisons in linkage disequilibrium and Hardy–Weinberg tests by calculating the P values adjusted for FDR (False Discovery Rate) with the R function fdrtool for R v2.13 (R Core Development Team 2013).
Because siblings could remain in the samples despite our cautious sampling procedure, we used COLONY v2.0 (Wang 2004) within each sampled pond to completely avoid sib-effect. For each pair, COLONY computed the probability of being full-sibs and half-sibs, which allows excluding related individuals for further analyses. Following Wang (2004)'s recommendations, each analysis was performed three times using the same information, with different seed numbers in order to avoid convergence problems commonly observed with maximum-likelihood estimation. We obtained identical family structures in all cases.
For each site, we calculated the mean numbers of alleles per locus (Na), numbers of effective alleles (Ne), allele frequencies, and observed (H o ) and expected (H e ) heterozygosities using GENALEX v6.5 (Peakall and Smouse 2005). F IS , and its statistical significance (Weir and Cockerham 1984) were calculated in FSTAT v2.9 (Goudet 1995).
Analysis of the genetic structure of the two species
We first assessed the genetic structure of our dataset, in order to verify the relevance of performing the genetic diversity analyses at the scale of each site. We estimated global F ST between ponds and their statistical significance (Weir and Cockerham 1984) with FSTAT v2.9 (Goudet 1995). We carried out Bayesian clustering analyses implemented in STRUCTURE v2.3 (Pritchard et al. 2000). Because the assignment of individuals has been shown to be sensitive to small sample sizes (Evanno et al. 2005), we excluded ponds with less than 10 individuals from those analyses. For each data set, we performed 10 replicate runs with a burn-in of 150,000 Markov chain Monte Carlo (MCMC) iterations followed by 106 iterations for values of K ranging from 1 to 10. We used the admixture model with correlated allele frequencies and the locprior option (Hubisz et al. 2009). This model uses the sampling location information to constrain the prior distribution for each assignment of individuals. The locprior option tends to outperform classical models when the actual structure is weak, and shows robustness to misdetection of wrong structures correlated with sampling locations (Hubisz et al. 2009; Hoffman et al. 2011; Moura et al. 2012). We determined the most likely values of K using the Evanno et al. (2005) method implemented in the program STRUCTURE HARVESTER (Earl and vonHoldt 2012). We used CLUMPP v1.1 (Jakobsson and Rosenberg 2007) to find optimal alignments of our 10 replicate cluster analyses. We finally used DISTRUCT v1.1 (Rosenberg 2004) for cluster visualization.
Effect of landscape composition on genetic diversity
We investigated which landscape features-with a special emphasis on quarries-affect the genetic diversity of the two study species. We assessed genetic diversity through the allelic richness within each pond as implemented in the ARES package (Van Loon et al. 2007) implemented in R v2.6 (R Core Team 2013). ARES counteracts the bias due to the strong dependence between genetic diversity and sample size by estimating allelic richness from accumulation curves. The curves estimate the expected number of unique alleles in a population for a given sample of individuals with 95 % confidence bounds. This method is more powerful than the intensively used rarefaction approach because it uses all valuable information from the original genetic data set (Van Loon et al. 2007). The number of bootstrap runs performed to calculate confidence bounds was set to 200. The accumulation curves were calculated for a maximum number of individuals of 100. After visual inspection of the graphs, sample size was set to n = 15 for B. bufo and n = 6 for B. calamita, which allowed us to extrapolate estimates before the curve asymptotes as recommended by Colwell et al. (2004).
We performed spatial autocorrelation analyses to assess the spatial scale at which landscape effects on genetic diversity had to be investigated. Such analyses assess the degree of genetic similarity among individuals separated by varying geographic distances. Significant autocorrelation at a distance class means that individuals separated by distances included in that class are more genetically similar than expected by chance. Visual inspection of correlograms (graphs of autocorrelation coefficients for the different distance classes) hence allows the detection of the spatial scale at which gene flow occurs, and hence the most appropriate scale for the analysis of small-scale landscape effects on genetic structure. We performed these analyses with GENALEX v6.5 (Peakall and Smouse 2005) with 1 km distance classes (the finest scale with sufficient sample size). We performed tests for statistical significance using 1,000 random permutations and bootstrap estimates.
Next, we estimated the area of 18 different classes of land cover within 2 km buffers around each pond (the buffer size was based on the results of the autocorrelation analysis, see Results) (Table 1). Those classes represented the major natural and anthropogenic features composing the study landscapes, and especially quarries. We estimated the total area of quarries and of the different types of elements within their perimeter of activities to test both the global effect of quarrying and the particular effect of each type of rehabilitated land. Because the spatial configuration of ponds may strongly influence effective dispersal in amphibians (e.g., Arens et al. 2007), we also assessed a connectivity index of ponds. We used CONEFOR SENSINODE v2.6 (Saura and Pascual-Hortal 2007a, 2007b) and its ArcGIS extension CONEFOR INPUTS to compute the Integral Index of Connectivity (IIC) of all potential breeding ponds within each buffer zone. The IIC is a graph-based index that takes into account patch area and the topological distance between them (Pascual-Hortal and Saura 2006). This index was only computed for B. bufo since B. calamita mainly breeds in small ephemeral ponds which configuration varies widely across time (Beebee 1983), making the inference of their locations from aerial photographs very difficult and irrelevant. We used a threshold dispersal distance of 2 km based on autocorrelation analysis.
To account for potential delays in landscape changes and their effects on genetic structure, we estimated landscape composition around the ponds at three distinct time points separated by ca. 10 generations for both species: mid 1940s (1946–1949), 1970s (1969–1974) and 2000s (2003–2013) (the years varied slightly among sites depending on land cover data availability, see supplementary material S1). To estimate their area within the buffers around ponds, we digitized landscape features by visual analysis based on black and white aerial photographs ranging from 1:25,000 to 1:30,000 scale and a resolution of 40 cm for the first two time points (IGN, BD Prises de vues aériennes ©), and real-colour geographically corrected aerial photographs at a 1:5,000 scale and a resolution of 50 cm for the most recent one (IGN, BD Ortho ©). We excluded opposite riverfronts from our sites in the Loire and Seine valleys since large rivers can be considered as a complete barrier to toad movements (Janin et al. 2009).
We used linear mixed effect models to quantify the contribution of quarrying and of the rest of the surrounding landscape on genetic diversity of both B. bufo and B. calamita. Mixed effect models allow analysis of nested and pseudo-replicated data such as those from sample sites exhibiting spatial autocorrelation. In our case, allelic richness in ponds from the same site are expected to be closer to each other than they are with ponds from other sites (i.e., spatial pseudo-replication). Instead of building separate models for each site, mixed effects models allow simultaneous analysis of the whole data set that incorporates the spatial autocorrelation. We treated the areas of the 18 land cover classes and the connectivity index as fixed factors and the sampling location as a random factor. Following Zuur et al. (2009), we used top-down model selection with Maximum Likelihood based AIC model comparison and Restricted Maximum Likelihood model validation. Log transformation and square-root transformation of the explanatory factors were used when necessary. Final validation was based on visual observation of the model residuals. All models were fitted using the nlme package (Pinheiro and Bates 2000) implemented in R v15.3.0 (R Core Team 2013).
Results
Data quality
In total, 1083 B. bufo and 957 B. calamita were sampled. Due to the lack of breeding ponds, the V area was not successfully sampled for B. bufo nor the G and L areas for B. calamita. COLONY analyses resulted in the removal of 101 sibs for B. bufo and 222 sibs for B. calamita. Our final data set was hence made of the genotypes of 982 B. bufo individuals and 735 B. calamita individuals, distributed in respectively 51 and 39 ponds, with sample sizes ranging from 5 to 41 individuals per pond, and averaging 19 individuals per pond for both B. bufo and B. calamita (Supplementary material S3). Within sites, distances between nearest neighbour ponds ranged from 0.3 to 5.2 km for B. bufo, and from 0.5 to 8.4 km for B. calamita; the maximum distance between ponds from the same site was 41.6 km for B. bufo and 43.4 km for B. calamita.
We found strong evidence for null alleles at four loci for B. bufo (Bufo_15, Bufo_39, Bufo_62, Bufo_65) concerning four of our sites and two loci for B. calamita (Cala_12, Cala_3a) for all sites. After re-amplification of all homozygotes, only loci Bufo_15 for B. bufo and Cala_12 and Cala_3a for B. calamita showed evidence for null alleles in more than two sites. These loci were removed from the analyses.
No pair of loci showed significant linkage disequilibrium after correction of P values for multiple comparisons (all P > 0.05, results not shown). Significant deviations from Hardy–Weinberg equilibrium were found in B. bufo for Bbufμ49 and Bbufμ65 in the B area, and for both species over all loci in each area, except in A for B. bufo (results not shown). This disequilibrium is most likely due to Walhund effects due to substructure in the dataset (as shown below) and a moderate frequency of null alleles which remained for Bbufμ65 in the B area (frequency = 0.060).
In the end, 10 and 11 loci were retained respectively for B.bufo and B. calamita. The mean number of alleles per locus across sample sites ranged respectively from 13.4 to 18.2 for B. bufo and from 5.9 to 8.9 for B. calamita (Table 2).
Analysis of the genetic structure of the two species
F ST levels were similar in the two species except in site P, where genetic differentiation was particularly high (F ST = 0.135) for B. calamita but remained low for B. bufo (F ST = 0.025) (Table 2).
STRUCTURE analyses revealed two clusters in the data set (B. bufo: ΔK2 = 88.96; B. calamita: ΔK2 = 811.11; Supplementary material S4), matching the limits of the main river drainages: A, G, L and P for the Seine, and B and V for the Loire (not shown). A second level of structure was detected, where each cluster matched the sites (five clusters for B. bufo and four for B. calamita) (Supplementary material S4). We therefore considered our sites as effective genetic units for further analyses.
Effect of landscape composition on genetic diversity
The corrected allelic richness per pond averaged per sample site ranged respectively from 73.5 to 82.6 for B. bufo and from 36.1 to 40.9 for B. calamita (Table 2). For both species, the sites located throughout the Seine valley (i.e., A, G, P) showed the lowest number of alleles (Table 2).
Autocorrelograms revealed significant spatial correlation among genotypes for geographic distances less than 2 km for both species in all but 1 site (Fig. 2): in P, r-values remained positively significant up to 3 km for B. bufo and 4 km for B. calamita. Based on those results, we chose a buffer size of 2 km for the genetic diversity analyses.
Correlograms showing genetic similarity among individuals (r) in relation to the distance among them, based on a distance class size of 1 km, for B. bufo and B. calamita, in each site. Dashed lines represent 95 % confidence intervals of the null hypothesis (r = 0) and error bars represent the 95 % confidence limits of r
For B. bufo, genetic diversity was best explained by landscape at the oldest time point. The best model, based on the 1940s landscape, included three landscape features: the area of dense woods and herbaceous vegetation, both positively correlated with allelic richness, and the area of roads, negatively correlated with allelic richness (Table 3 and Fig. 3). For the 1970s landscape, model selection retained three landscape features as well: the surfaces area of semi-wooded areas and of herbaceous areas showed positive relationship with allelic richness whereas urbanized areas were negatively correlated. The final model for the 2000s landscapes only included wetland areas, a factor that was not significant in the 1940s and 1970s models. The connectivity index was not retained in any of the three models, as well as for all within-quarry habitats.
Relationship between allelic richness and the areas of land cover (square-rooted ha) that were selected in linear mixed models, for B. bufo (1970s) and B. calamita (2000s). Landscape variables were square rooted. Each site is represented with a distinct symbol (A: dark square, B: diamond; G: white square; L: circle; P: cross and V: circle). Lines: fitted values for each site
For B. calamita, the 2000s landscape best explained its current genetic diversity. The final model included three landscape features: the area of dense woods, negatively correlated with allelic richness, the areas of bare ground and the total areas of quarries, both positively correlated with allelic richness (Table 3 and Fig. 3). Off-quarry bare ground was marginally non-significant in the 2000s model, though. The models for 1940s and 1970s included the same variables, except for the area of dense woodlands, which was not kept in the 1970s model. Similarly to B. bufo, no correlation were found for all within-quarry habitats
For both species and for each three time points, graphs depicting the significant relationships between land cover and genetic diversity are presented in Supplementary Material S5.
Discussion
Genetic diversity mostly depends on effective population size and incoming gene flow (Frankham et al. 2010). Focusing on genetic diversity hence provides a good diagnostic of environmental quality (through its effects on population sizes) and landscape connectivity (through its effects on gene flow).
Different landscape features affect B. bufo and B. calamita
Our results matched well the known habitat preferences of B. bufo. In this species, population connectivity is usually thought to be positively affected by forest cover and herbaceous land and negatively affected by mineralized areas. Indeed, the first two elements have been recognized as the species habitats for home-ranging and dispersal (Denton 1991; Denton and Beebee 1994; Scribner et al. 2001; Hartel et al. 2008; Janin et al. 2009), whereas high proportions of artificial land have been known to act as a barrier to its movements (Hitchings and Beebee 1998). In our analyses, a positive influence of dense and semi-dense woods was found in the 1940s and 1970s models, respectively. A negative influence of mineralized areas was detected through the effect of roads in the 1940s model and the effect of urbanization in the 1970s model. In addition to resistance to movements of the mineralized areas, roads and urban areas support car traffic that could cause dramatic B. bufo mortality during dispersal and seasonal migration (Beebee 2013). Although roads with low traffic are excellent hunting places, with warmer temperatures at night than in the surroundings, only their negative effect predominated in our results.
While wetlands have been observed to sometimes enhance the connectivity among B. bufo populations (Piha et al. 2007), the statistical signal of that landscape element in the 2000s models is probably an artefact rather than a truly influential factor. In particular for the site L, the two ponds L3 and L21 are suspected to have an overproportional effect in the models because their high genetic diversity values stand out from the others (Supplementary Material S5). The leverage calculation for both ponds confirmed the suspected artefact (i.e., for i = 1, n populations, h i > 4/n). This would explain why wetlands appeared as significant when they were the only factor in the model, as was the case for the 2000s models (wetlands were also significant if they were the only factor in the 1940s and 1970s model (1940s: p = 0.0166, AIC = 299.86; 1970s: p = 0.0039, AIC = 297.14) but were not retained in the final 1970s model).
For B. bufo, we did not find an effect of pond configuration at any time point. The patches considered in the configuration analysis include many quarrying lakes. Although such “super-ponds” support 65 % of the sampled ponds for B. bufo, they form large and deep contiguous structures (e.g., up to 322 ha in one piece) that may act as barriers for poor swimmers such as B. bufo (Denton 1991). Moreover, total quarrying areas showed no significant influence on genetic diversity in B. bufo, despite the fact that they account for both high vehicle traffic and areas of bare ground.
Our results also matched our expectations based on the biology of B. calamita: genetic diversity was negatively affected by the area of dense woods, and positively affected by bare grounds and quarries. It is interesting to note that, unlike for B. bufo, the same landscape variables-except for dense woodlands-were retained in the final models of the 1940s, 1970s and 2000s. This could be due to the fact that those landscape elements were spatially correlated over time. Bare ground soil was especially large and influential in the B site (Fig. 3), where alluvial deposits of the Loire river create large sandy habitats restricted to the same river loops. Similarly, quarrying has spread around historic sites and was restricted to sand and gravel placers.
Our study sheds light on the influence of quarries on fragmentation and biodiversity maintenance. Quarries showed no deleterious effects on B. bufo genetic diversity. This may be because (1) the quarrying area was relatively small in the 1940s and 1970s (i.e., mean quarrying area within 2 km buffers < 5 ha in the 1940s and < 65 ha in the 1970s), and as a result the land conversion impact of quarrying may have been too limited to be detected at these time points. Alternatively, (2) quarries may have provided suitable habitats, favouring both demographic and gene flow processes, that would have compensated for the potential negative effects of bare soil or wide contiguous lakes that are found within quarries. Indeed, in 1940s and 1970s the amount of open and mineralized areas, which are unsuitable for B. bufo, represented less than 51 % of the total quarry areas, while the rest consisted of a mosaic of habitats suitable for home-ranging and dispersal (e.g. wetlands, grassland, shrubby and dense woodlands).
In the case of B. calamita, quarries were positively correlated with genetic diversity in all models. This supports our hypothesis that quarries can constitute pioneer environments that favour B. calamita, in particular through enhanced home-ranging and breeding (Kovacs 2001; Voetzel et al. 2008). Unfortunately, we were unable to identify the type of within-quarry habitat that enhances B. calamita genetic diversity. This may be due to the quick turnover of land use inside quarries. Indeed, in our sites the combined processes of extraction and rehabilitation often caused more than 10 ha/year of land use transfer, which accounts for 10 % to 100 % or more of the original quarry. Consequently, the three time points we used did not allow us to capture the dynamics of land use change, and therefore identify all effects associated with landscape composition within quarries.
Temporal scale of landscape effects on genetic diversity
Our analyses showed that the 1940s landscape best explained B. bufo’s genetic diversity whereas the 2000s landscape was the most predictive for B. calamita. In all study sites, landscape changes were stronger between the last two time points (i.e., 1970s–2000s). For example, the area of herbaceous vegetation surrounding ponds was reduced by 7 % (8 ha) between the 1940s and the 1970s, and by 21 % (i.e., 85 ha) between the 1970s and the 2000s. Urban areas increased at similar rates for both time intervals, i.e., by 49 % (i.e., 37 ha) between the 1970s and the 2000s and by 44 % (i.e., 20 ha) between the 1940s and the 1970s, but newly urbanized surface areas were doubled between the last two time points. In the same way, quarrying was much more intense during the most recent time period (i.e., 4 890 ha excavated) rather the oldest one (i.e., 2 130 ha excavated). Hence, the most recent and intense landscape change was shown to match best the genetic diversity of B. calamita whereas that of B. bufo was best explained by the oldest landscape. Two non-exclusive hypotheses can explain this difference.
First, landscape dynamics have affected differently the preferential dispersal habitats of the two species. Since the 1940s, herbaceous vegetation has been constantly reduced whereas urban areas have increased, two drivers which probably resulted in a loss of connectivity for B. bufo. On the contrary, B. calamita may have benefited from a global gain of connectivity since quarrying increased dramatically throughout the two time periods. Parallel to this, it may take dozens of generations to reach equilibrium between genetic drift and gene flow after a fragmentation event, whereas only one generation is sufficient to detect migrants (Crow and Aoki 1984; Varvio et al. 1986; Frankham et al. 2010). Thus, the temporal discrepancy between our focal species could be linked to the global evolution of the landscape in regards to their preferential dispersal habitats.
Second, genetic drift operates more slowly in large populations than in small ones (Frankham et al, 2010). B. bufo being more abundant than B. calamita in our study sites, its genetic structure may therefore respond more slowly to changes in gene flow and demography due to landscape changes (e.g. Gauffre et al. 2008). Under that second potential explanation, our results would suggest that the landscape changed faster than genetic composition in B. bufo between the last two time points (i.e., 1970s–2000s).
Again, these conclusions underline the importance of time scale in landscape genetic studies, as previously highlighted by several authors before (e.g., Orsini et al. 2008; Epps et al. 2013).
Quarries can counteract natural habitat losses for pioneer species
We showed here that quarrying can favour the genetic diversity of a pioneer species, B. calamita, and therefore may positively support the viability of its populations. This raises the potential role of quarries as substitution zones for home-ranging, breeding and dispersal habitat for this species. This role may become essential in areas where natural habitats have largely been replaced, such as valleys with channelized riverbeds and intensive anthropogenic land use. For example, this may be the case in the Seine basin: quarries support 71 % of existing breeding sites of B. calamita while natural habitat only accounts for 11 %. Our results further support the idea that quarries may also be beneficial in less disturbed sites such as the site B from the Loire basin (Fig. 3). Hence, quarries do not only represent a “second-choice” for B. calamita but may also play a role when natural habitat is available. Interestingly, breeding of B. calamita in quarries mainly occurs in active zones despite additional mortality of juveniles and adults due to truck movement, stock transfer, dust emission and aggregate extraction. Indeed, active sites represented 70 % of the ponds in which we were able to collect B. calamita, and support large populations that contribute to gene flow. The extreme conditions encountered in operating quarries may drastically reduce competition and predation risks. In particular, they would be sufficient to stop colonization by competitors such as B. bufo (Denton and Beebee 1994). However, extractive activity could generate strong selection pressures on B. calamita that would necessitate its adaptation. The positive effect of quarrying on B. calamita genetic diversity may facilitate such adaptation, mitigating demographic decline. Large genetic diversity would favour evolutionary rescue and thus population survival (Gonzalez et al. 2012).
Similar to B. calamita, numerous endangered species have been observed to seek refuge in active quarries (e.g., plants: Lythrum thymifolium; invertebrates: Oxygastra curtisii; reptiles: Lacerta lepida; amphibians: Bombina variegata; birds: Burhinus oedicnemus; (Voetzel et al. 2008)). These are generally ruderal species occurring in early successional stages, and competitively weak species living in nutrient-poor habitats. Quarries, by providing good-quality habitat and, sometimes, favouring genetic diversity, may support the long-term conservation of pioneer species. However, conservation strategies also have to focus on other groups of species likely to be negatively impacted by quarrying. Our results based on B. bufo genetic diversity suggest that this impact could be mitigated if habitats of different nature were maintained at intermediate proportions. This paradox advocates for further research to understand the consequences of quarrying effects on community composition, dynamics and evolution.
Conclusion
Our study showed that landscape composition affected genetic diversity in B. bufo and B. calamita. Landscape change seemed to operate at different time scales on these species, confirming that the reconstruction of landscape history is essential before any inference. Our results suggest new insights on genetic conservation and long-term management of quarries. No effect of quarrying was found for B. bufo, presumably due to the joint effect of a time lag between landscape and genetic processes and of antagonistic effects of within-quarry habitats. However, we found a positive effect of quarry sites on the genetic diversity of B. calamita, suggesting that this activity may contribute to B. calamita’s conservation despite heavy anthropogenic disturbances. From a broader perspective, long-term conservation strategies of endangered pioneer species should take into account the positive influence of this industrial activity, for example when deciding the type of rehabilitation to be applied to quarry areas after exploitation.
References
Arens P, van der Sluis T, van’t Westende WPC, Vosman B, Vos CC, Smulders MJM (2007) Genetic population differentiation and connectivity among fragmented Moor frog (Rana arvalis) populations in The Netherlands. Land Ecol 22:1489–1500
Banks B, Beebee TJC, Cooke AS (1994) Conservation of the Natterjack toad Bufo calamita in Britain over the period 1970–1990 in relation to site protection and other factors. Biol Cons 67:111–118
Barnaud G, Le Bloch F (1998) Entre Terre et Eau, Agir pour les zones humides, Dossier d’information. Ministère de l’aménagement du territoire et de l’environnement, Paris, France
Beebee TJC (1983) The Natterjack Toad. Oxford University Press, Oxford
Beebee TJC (2013) Effects of road mortality and mitigation measures on amphibian populations. Cons Biol 27:657–668
Belkhir K et al (1996–2004) GENETIX 4.05, logiciel sous Windows TM pour la Génetique des populations. Laboratoire genome, populations, interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France. URL: http://www.genetix.univmontp2.fr/genetix/intro.htm
Benes J, Kepka P, Konvicka M (2003) Limestone quarries as refuges for European xerophilous butterflies. Cons Biol 17:1058–1069
Berhe AA (2007) The contribution of landmines to land degradation. Land Degrad Dev 18:1–15
Blanchong JA, Sorin AB, Scribner KT (2013) Genetic diversity and population structure in urban white-tailed deer. J Wild Manag 77:855–862
Brandle M, Durka W, Altmoos M (2000) Diversity of area dwelling beetle assemblages in open-cast lignite mines in Central Germany. Biodivers Cons 9:1297–1311
Brede EG, Rowe G, Trojanowski J, Beebee TJC (2001) Polymerase chain reaction primers for microsatellite loci in the common toad Bufo bufo. Mol Ecol Notes 1:308–310
Bzdon G (2008) Gravel pits as habitat islands: floristic diversity and vegetation analysis. Pol J Ecol 56:239–250
Carlsson J, Mcdowell JR, Diaz-Jaimes P, Carlsson JEL, Boles SB, Gold JR, Graves JE (2004) Microsatellite and mitochondrial DNA analyses of Atlantic bluefin tuna (Thunnus thynnus thynnus) population structure in the Mediterranean Sea. Mol Ecol 13:3345–3356
Clements R, Sodhi NS, Schiltuizen M, Kling P (2006) Limestone Karsts of Southeast Asia: imperiled arks of biodiversity. BioSci 56:733–742
Colwell RK, Mao CX, Chang J (2004) Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecol 85:2717–2727
Cooke AS, Oldham RS (1995) Establishment of populations of the common frog, Rana temporaria, and common toad, Bufo bufo, in a newly created reserve following translocation. Herpetol 5:173–180
Crow JF, Aoki K (1984) Group selection for a polygenic behavioural trait—estimating the degree of population subdivision. Proc of the Natl Acad of Sci of the USA Biol Sci 81:6073–6077
Cushman SA (2006) Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol Cons 128:231–240
Denton JS (1991) The terrestrial ecology of the natterjack Bufo calamita and the common toad, Bufo bufo. Dissertation, University of Sussex
Denton J, Beebee TJC (1991) Terrestrial ecology of the natterjack toad Bufo calamita. In: Korsos Z, Kiss I (eds) Proceedings of the Sixth Ordinary General Meeting. S. E. H, Budapest, pp 137–141
Denton JS, Beebee TJC (1993) Reproductive strategies in a female biased population of Natterjack toads, Bufo calamita. Anim Behav 46:1169–1175
Denton JS, Beebee T (1994) The basis of niche separation during terrestrial life between two species of toad Bufo bufo and Bufo calamita: competition or specialisation? Oecol 97:390–398
Denton JS, Beebee TJC (1996) Double clutching by natterjack toads Bufo calamita at a site in southern England. Amphib Reptil 17:159–167
Dixo M, Metzger JP, Morgante JS, Zamudio KR (2009) Habitat fragmentation reduces genetic diversity and connectivity among toad populations in the Brazilian Atlantic Coastal Forest. Biol Cons 142:1560–1569
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Cons Genet Resour 4:359–361
Eggert C, Miaud C (2004) Estimation de la réussite du déplacement de populations de Crapaud calamite et Pélodyte ponctué dans le cadre de l’aménagement du Port Autonome du Havre Port 2000-Bilan au terme de la 3ème année. LBPA Université de Savoie, Le Bourget du Lac
Epps CW, Palsoll PJ, Wehausen JD, Roderick GK, Ramey RR II, McCullough DR (2005) Highways block gene flow and cause a rapid decline in genetic diversity of desert bighorn sheep. Ecol Lett 8:1029–1038
Epps CW, Wasser SK, Keim JL, Mutayoba BM, Brashares JS (2013) Quantifying past and present connectivity illuminates a rapidly changing landscape for the African elephant. Mol Ecol 22:1574–1588
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Evol Syst 34:487–515
Frankham R, Balou J, Briscoe D (2010) Introduction to Conservation Genetics. Cambridge University Press, Cambridge
Gauffre B, Estoup A, Bretagnolle V, Cosson JF (2008) Spatial genetic structure of a small rodent in a heterogeneous landscape. Mol Ecol 17:4619–4629. doi:10.1111/j.1365-294X.2008.03950.x
Glandt D (1986) Die saisonalen wanderungen der mitteleuropaischer amphibicn. Bonn Zool Beitr 37:211–228
Goldberg CS, Waits LP (2010) Comparative landscape genetics of two pond-breeding amphibian species in a highly modified agricultural landscape. Mol Ecol 19:3650–3663
Gonzalez A, Ronce O, Ferriere R, Hochberg ME (2012) Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Phil Trans of R Soc- Biol Sci 368:1610
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Halley JW, Oldham RS, Arntzen JW (1996) Predicting the persistence of amphibian populations with the help of a spatial model. J Appl Ecol 33:455–470
Hanski I (1999) Metapopulation ecology. Oxford University Press, Oxford
Hartel T, Nemes S, Demeter L, Oellerer K (2008) Pond and landscape characteristics - which is more important for common toads (Bufo bufo)? A case study from central Romania. Appl Herpetol 5:1–12
Hitchings SP, Beebee JTC (1998) Loss of genetic diversity and fitness in Common Toad (Bufo bufo) populations isolated by inimical habitat. J Evol Biol 11:269–283
Hof C, Araujo MB, Jetz W, Rahbek C (2011) Additive threats from pathogens, climate and land-use change for global amphibian diversity. Nat 480:516–519
Hoffman JI, Peck LS, Linse K, Clarke A (2011) Strong population genetic structure in a broadcast-spawning antarctic marine invertebrate. J Hered 102:55–66
Holzhauer SIJ, Ekschmitt K, Sander A, Dauber J, Wolters V (2006) Effect of historic landscape change on the genetic structure of the bush-cricket Metrioptera roeseli. Landsc Ecol 21:891–899
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Hughes A et al (2008) Ecological consequences of genetic diversity. Ecol Lett 11:609–623
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinforma 23:1801–1806
Janin A, Léna JP, Ray N, Delacourt D, Allemand P, Joly P (2009) Assessing landscape connectivity with calibrated cost-distance modelling: predicting common toad distribution in a context of spreading agriculture. J App Ecol 46:833–841
Karraker N, Gibbs JP (2009) Amphibian production in forested landscapes in relation to wetland hydroperiod: a case study of vernal pools and beaver ponds. Biol Cons 142:2293–2302
Kovacs JC (2001) Le patrimoine écologique des zones humides issues de l’exploitation des carrières. Etude Ecosphère- Comité Nationale de la Charte Environnement de l’UNPG
Lameed GA, Ayodele AE (2010) Effect of quarrying activity on biodiversity: case study of Ogbere site, Ogun State Nigeria. African J Env Sci Technol 4:740–750
Leimu R, Mutikainen P, Koricheva J, Fischer M (2006) How general are positive relationships between plant population size, fitness and genetic variation? J Ecol 94:942–952
Martino L, Fritz M (2008) New insight into land cover and land use in Europe. Eurostat 33/2008
Moura AE, Natoli A, Rogan E, Hoelzel AR (2012) Atypical panmixia in a European dolphin species (Delphinus delphis): implications for the evolution of diversity across oceanic boundaries. J Evol Biol 26:63–75
Novak J, Konvicka M (2006) Proximity of valuable habitats affects succession patterns in abandoned quarries. Ecol Eng 26:113–122
Orsini L, Corander J, Alasentie A, Hanski I (2008) Genetic spatial structure in a butterfly metapopulations correlates better with past than present demographic structure. Mol Ecol 17:2629–2642
Pascual-Hortal L, Saura S (2006) Comparison and development of new graph-based landscape connectivity indices: towards the priorization of habitat patches and corridors for conservation. Landsc Ecol 21:959–967
Peakall R, Smouse P (2005) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE, Huff DR (1995) Evolutionary implications of allozyme and RAPD variation in diploid populations of buffalograss Buchloe dactyloides. Mol Ecol 4:135–147
Peakall R, Ruibal M, Lindenmayer DB (2003) Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evol 57:1182–1195
Piha H, Luoto M, Merila J (2007) Amphibian occurrence is influenced by current and historic landscape characteristics. Ecol Appl 17:2298–2309
Pinheiro JC, Bates DM (2000) Mixed-effects models in S and S-plus. Springer, London
Pope SE, Fahrig L, Merriam NG (2000) Landscape complementation and metapopulation effects on leopard frog populations. Ecol 81:2498–2508
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genet 155:945–959
Rannap R, Lohmus A, Jakobson K (2007) Consequences of coastal meadow degradation: the case of the natterjack toad (Bufo calamita) in Estonia. Wetl 27:390–398
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reading CJ, Loman J, Madsen T (1991) Breeding pond fidelity in the common toad, Bufo bufo. J Zoo Soc Lond 225:201–211
Rogell B, Gyllenstrand N, Höglund J (2005) Six polymorphic microsatellite loci in the natterjack toad Bufo calamita. Mol Ecol Notes 5:639–640
Romero J, Real R (1996) Macroenvironmental factors as ultimate determinants of distribution of common toad and natterjack toad in the south of Spain. Ecogr 19:305–312
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genet 145:1219–1228
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Rowe G, Beebee TJC, Burke T (1997) PCR primers for polymorphic microsatellite loci in the anuran amphibian Bufo calamita. Mol Ecol 6:401–402
Rowe G, Beebee TJC, Burke T (2000) A microsatellite analysis of natterjack toad, Bufo calamita, metapopulations. Oikos 88:641–651
Rowe G, Beebee TJC, Burke T (2001) A further four polymorphic microsatellite loci in the natterjack toad Bufo calamita. Conserv Genet 1:371–372
Santoul F (2000) L’avifaune aquatique des gravières de la plaine alluviale de la Garonne Dissertation, University of Toulse III
Santoul F, Gaujard A, Angélibert S, Mastrorillo S, Céréghino R (2009) Gravel pits support waterbird diversity in an urban landscape. Hydrobiol 634:107–114
Saura S, Pascual-Hortal L (2007a) Conefor Sensinode 2.2. Users manual. Software for quantifying the importance of habitat patches for landscape connectivity through graphs and habitat availability indices, University of Lleida, Spain
Saura S, Pascual-Hortal L (2007b) A new habitat availability index to integrate connectivity in landscape conservation planning: comparison with existing indices and application to a case study. Landsc Urban Plan 83(2–3):91–103
Schlupp I, Podloucky R (1994) Changes in breeding site fidelity: a combined study of conservation and behaviour in the common toad Bufo bufo. Biol Cons 69:285–291
Schulz F, Wiegleb G (2000) Development options of natural habitats in a post-mining landscape. Land Degrad Dev 11:99–110
Scribner KT, Arntzen JW, Cruddace N, Oldham RS, Burke T (2001) Environmental correlates of toad abundance and population genetic diversity. Biol Cons 98:201–210
Sinsch U (1989) Migratory behaviour of the common toad (Bufo bufo) and the natterjack toad (Bufo calamita). In: T.E.S. Langton (ed) Amphibians and Roads. pp 113–125
Sinsch U (1990) Migration and orientation in anuran amphibians. Ethol Ecol Evol 2:65–79
Sinsch U (1992) Structure and dynamic of a Natterjack toad metapopulation (Bufo calamita). Oecol 90:489–499
Sinsch U (1997) Postmetamorphic dispersal and recruitment of first breeders in a Bufo calamita metapopulation. Oecol 112:42–47
Sinsch U, Oromi N, Miaud C, Denton J, Sanuy D (2012) Connectivity of local amphibian populations: modelling the migratory capacity of radiotracked natterjack toads. Anim Cons 15:388–396
Smith MA, Green DM (2005) Dispersal and the metapopulation paradigm in amphibian ecology and conservation: are all amphibian populations metapopulations? Ecogr 28:110–128
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Hered 82:561–573
Stevens VM, Polus E, Wesselingh RA, Schtickzelle N, Baguette M (2004) Quantifying functional connectivity: experimental evidence for patch-specific resistance in the natterjack toad (Bufo calamita). Landsc Ecol 19:829–842
Stevens VM, Verkenne C, Vandewoestijne S, Wesselingh RA, Baguette M (2006) Gene flow and functional connectivity in the natterjack toad. Mol Ecol 15:2333–2344
Taylor PD et al (1993) Connectivity is a vital element of landscape structure. Oikos 68:571–573
R Core Team (2013). R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/
Tropek R, Konvicka M (2008) Can quarries supplement rare xeric habitats in a piedmont region? Spiders of the Blanksi les Mts, Czech Republic. Land Degrad Develop 19:104–114
Van Loon EE, Cleary DFR, Fauvelot C (2007) ARES: software to compare allelic richness between uneven samples. Mol Ecol Notes 7:579–582
Van Oosterhout C, Hutchinson WF, Willis DPM, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Varvio SL, Chakraborty R, Nei M (1986) Genetic-variation in subdivided populations and conservation genetics. Hered 57:189–198
Voetzel D, Rohaut M, Arbion M (2008) Potentialités écologiques des carrières de roche massive. Etude UNPG
Wang J (2004) Sibship reconstruction from genetic data with typing errors. Genet 166:1963–1979
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evol 38:1358–1370
Wilcox BA, Murphy DD (1985) Conservation strategy – The effects of fragmentation on extinction. Am Nat 125:879–887
Zellmer AJ, Knowles LL (2009) Disentangling the effects of historic vs. contemporary landscape structure on population genetic divergence. Mol Ecol 18:3593–3602
Zuur AF, Ieno EN, Walker NJ, Saveliev AA, Smith GM (2009) Mixed effects models and extensions in ecology with R. Statistics for Biology and Health. Springer-Verlag, New York
Acknowledgments
We are indebted to Bernard Frochot, Jean-Claude Lefeuvre, Pierre Joly, Stéphanie Manel, Steve Palmer, Virginie Stevens and Justin Travis for helpful comments on this work. We are also grateful to Khaldia Akkar and Marielle Perroz for technical hep during data collection, to Hugo Anest, Karen Cheurlot, Emeline Hudik, Basile Hurault, Jérémy Gauthier, Emilie Klam, Angeline Lesueur, Michael Pereira, Jennifer Thomas and Nicolas Zilbermann for field work, to Jeffrey Carbillet, Louise Keszler, Lise Lallemand, Jérôme Lin, Flore Loyer and Solène Sacré for GIS work, and to Florian Lesage, Josie Lambourdière and Jose Utge for genetic data collection. All molecular analyses were supported by the ‘Service de Systématique Moléculaire’ of the Muséum National d’Histoire Naturelle (UMS 2700; OMSI). We thank Julio Pedraza-Acosta (UMS 2700; OMSI) for providing high-performance computing facilities. We finally thank the French National Union of Aggregates and the participating quarrying companies which have led us to sample operating sites in the person of Yves Adam and Christian Béranger, and the engineering office ENCEM in the person of Olivier Verdier, Johan Gourvil and Pascal Maurel for technical information about quarry ecology. This study was co-funded by the French Agence Nationale de la Recherche et de la Technologie (ANRT) and the Engineering office ENCEM (Cifre Contract n° 1209/2010).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Flavenot, T., Fellous, S., Abdelkrim, J. et al. Impact of quarrying on genetic diversity: an approach across landscapes and over time. Conserv Genet 16, 181–194 (2015). https://doi.org/10.1007/s10592-014-0650-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0650-8