Abstract
Epithelial mesenchymal transition (EMT), as defined by loss of epithelial characteristics and gain of a mesenchymal phenotype, has been reported in vivo although the occurrence of events remains unclear. This study aims at exploration of EMT portraits of breast cancer (BC) with relevance to different molecular pathways, especially potential EMT triggers and BC molecular subtypes. Immunohistochemical (IHC) expression of markers/triggers of EMT was studied on a well-defined cohort of invasive non-lobular BC (n = 1,035), prepared as tissue microarrays. IHC panel of biomarkers included cadherins (cad; E-cad and N-cad), TGFβ1, PIK3CA, pAkt, and others. Reverse phase protein array (RPPA) was performed for quantitative analysis of proteins extracted from formalin fixed paraffin embedded tissues of a subset of cases from this cohort. Four combinatorial phenotypic groups representing cadherin switch were defined, including E-cad+/N-cad−, E-cad−/N-cad−, E-cad+/N-cad+, and E-cad−/N-cad+. Statistically significant association was noticed between these phenotypes and histological tumour grade, tumour type and size and NPI staging classes. The E-cad/N-cad switch occurred more frequently in the triple negative molecular class, both basal and non-basal, and in the HER2+ subtype than in luminal BC. Significant outcome differences were observed between cadherin switch combinatorial groups regarding BCSS and DMFS (p < 0.001). Results of RPPA confirm those observed using IHC regarding differential expressions of EMT markers/triggers. EMT/cadherin switch programs in BC appear to occur in synergy with TGFβ1 and PIK3/Akt pathways activation. These data explain, at translational proteomic level, the molecular heterogeneity and in turn the varied clinical behaviour of BC molecular subtypes. RPPA is a promising high-throughput technique in monitoring subtle quantitative changes in protein expression in archival material.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The term epithelial mesenchymal transition (EMT) describes a sequence of events during which epithelial cells lose many of their epithelial characteristics and gradually gain those typical of mesenchymal cells [1]. This phenomenon involves complex architectural and behavioural cellular changes encompassing a wide spectrum of inter-cellular and intra-cellular changes, which are probably determined by diverse extra-cellular signals [2]. Loss of epithelial characteristics and gain of mesenchymal characteristics confer invasive and migratory capacity to the malignant cells [3]. However, it is noteworthy that EMT does not necessarily denote a lineage switch; but rather a series of complex changes manifesting through phenotypic and functional alterations of malignant cells [1, 4]. EMT has been described with much emphasis in tissue culture and animal models using cancer cell lines [5, 6]. The manifestation of EMT in tissue cultures prompted some researchers to describe EMT as transient reversible process [3] that is a mere cell culture phenomenon which lacks direct clinical evidence or clear molecular markers in breast cancer [7].
Whether EMT is a prerequisite of tumour progression or not remains a controversial issue. For instance, EMT has been considered as a key role-player in cancer dissemination from local to remote sites [8] or at least as one possible mechanism explaining local invasiveness and progression of cancer [6, 9]. Giampieri et al. [10] demonstrated the impact of transient TGFβ signalling; an EMT trigger, in switching cancer cells from cohesive 'collective' to 'single cell' motility, which is essential for blood-borne metastasis. However, other authors [11] doubt its occurrence in real cancers and progression without EMT has been reported [12].
Insights into EMT triggering mechanisms indicate that several complex signalling pathways are involved in the initiation and execution of EMT in the contexts of cancer development and progression. These molecular mechanisms significantly overlap with those controlling cellular adhesion, motility, invasion, differentiation and survival [2, 13]. A number of specific molecular pathways and transcription factors have been reported as ‘'EMT triggers’' including TGFβ [14], Twist [15], PI3K/Akt [16] and CTEN [17]. When expressed in a variety of cell types, these factors act as transcriptional repressors of the epithelial adhesion molecule E-cadherin and alter the expression of a diverse number of genes denoting in-vitro EMT with subsequent promotion of cancer invasion and metastasis [17, 18].
Characteristic molecular phenotypic changes of carcinoma cells have been reported to accompany cancer progression through EMT program. For instance, down-regulation of E-cadherin and up-regulation of N-cadherin or 'cadherin switching', has been reported to enhance cellular motility of mammary epithelial cell lines [19]. Other molecular changes that reflect an EMT-like programs have been described in cancer cell lines including ovarian cancer cell lines [20], prostate cancer cell lines [21] and melanoma cells [22]. In the breast, EMT has been reported in some particular cancer molecular subtypes [23].
In view of the lack of exact identification of events associated with EMT in vivo and its clinical relevance in human breast cancer, the aim of this study was to elucidate the molecular changes that could trigger or be a reflection of EMT occurrence in early invasive breast cancer with relevance to the known breast cancer molecular subtypes and patients’ outcome. Relevant markers are assessed at the protein level using immunohistochemistry (IHC) and reverse phase protein array (RPPA).
RPPA represents a novel high-throughput technique that allows simultaneous measurement of protein expression levels in a large number of biological samples [24]. Minuscule amounts of proteins are individually spotted on a nitrocellulose-coated microarray slide which is incubated with a single specific primary antibody to detect expression of the target protein across many samples [25]. Therefore, depending on the design a single microarray can accommodate hundreds to thousands of samples that are printed in replicate. Detection and quantification of antigen antibody reaction is performed using either a primary or a secondary labelled antibody by chemiluminescent, fluorescent, or colorimetric assays [26].
Patients and methods
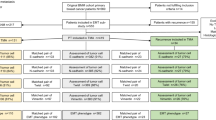
This study was conducted on a well-characterised series of early stage (pT1 and pT2) invasive non-lobular primary operable breast carcinoma (n = 1,035) from patients enrolled into the Nottingham Tenovus Primary Breast Carcinoma Series that presented at Nottingham City Hospital between 1990 and 1998. Patients were under the age of 70 years and managed in a uniform manner. Patients’ clinical history and tumour criteria, information on therapy and outcomes were available and prospectively maintained. Histological tumour types consisted of 723 (69.9 %) invasive ductal carcinomas no special type (NST), 196 (18.9 %) tubular mixed carcinomas, 54 (5.2 %) special types of excellent prognosis (tubular carcinomas, mucinous carcinomas, cribriform carcinomas and papillary carcinomas), 35 (3.4 %) medullary carcinomas and 27 (2.6 %) mixed NST and special type carcinomas. Data on a wide range of biomarkers of known clinical and biological relevance to breast cancer were available [27].
Outcome data included survival status, survival time in months, cause of death, recurrence free interval, time to loco-regional recurrence and/or distant metastasis. The breast cancer-specific survival (BCSS) is defined as the time (in months) from the date of primary surgery to the date of breast cancer-related death. DMFS is defined as the duration (in months) from the date of primary surgery to the appearance of distant metastasis. The patients had a median age at diagnosis of 54 years (range 18–70 years) with a median overall survival of 125 months (range 4–243 months) and the median time of event-free survival of 112 months (range 2-239 months). Distant recurrence occurred in 375 cases (31 %), while 313 (26 %) patients died from breast cancer, while 689 (59 %) patients were alive at the end of follow-up.
This study was approved by Nottingham Research Ethics Committee 2 under the title of 'Development of a molecular genetic classification of breast cancer'.
Immunohistochemistry
The tissue markers’ panel used in this study is formed of 19 markers of close relevance to breast cancer biology and prognosis in addition to those reflecting potential/proposed EMT triggers in breast or other cancers. With the exception of the anti-Ki67/MIB1 antibody which was assayed using full face paraffin embedded sections, these markers were assessed using TMA employing the standard Streptavidin–biotin complex method as previously described [28, 29]. Table 1 shows these markers and the rationale behind their inclusion in this study in addition to the antibodies sources and clones used.
Scoring of IHC stained markers
IHC stained TMA cores were scored following the semi-quantitative histochemical score (H-score) method which takes the intensity of staining and percentage of stained cells in consideration [33], giving a continuous measure (0–300) of marker expression within the invasive tumours. However, Ki-67/MIB1 labelling index (LI) was scored as the percentage of positively stained nuclei in 1,000 malignant cells on full face sections.
Reverse phase protein microarray
This study was conducted on 49 cases of primary invasive breast carcinoma patients selected out of a randomly selected subset from the Nottingham primary series (one-third of the cases included in this study, n = 345 case, chosen using SPSS random sampling approach). These 49 cases were primarily chosen based on the availability of paraffin blocks containing adequate tumourous tissue burden (see below) for protein extraction that could be dissected by macrodissection.
Invasive component dissection
From each block, one 4 μm section was cut and stained with haematoxylin and eosin (H&E) to check for block inclusion/exclusion and guide the process of macrodissection. For each block/case to be included, areas of invasive tumour had to constitute at least 50 % of the overall area of the tissue section. From each block, two 20 μm sections were cut and mounted on glass slides. Under light microscopy, the H&E sections were thoroughly scanned to determine and mark the spots of invasive breast carcinoma. Macrodissection was performed with the use of a disposable scalpel blade, with strict contamination-free handling conditions adopted throughout the procedure.
Protein extraction
Macro-dissected FFPE tissues representing each sample were transferred into a labelled eppendorf tube. Tissues were deparaffinised using 1 ml xylene (2 × 5 min), rehydrated in a graded ethanol series (100, 90, 70 %, 5 min each) and centrifuged to remove excess ethanol. A total of 150 μl of freshly prepared Laemmli buffer [34] were added and tubes sealed with plastic Parafilm prior to heating at 105 °C for 20 min. Samples were cooled for 5 min on ice, then centrifuged, where fluid supernatant pipetted into a new labelled sterile tube and stored at −80 °C.
Reverse phase protein microarray
4× SDS sample buffer was added to protein extracts in a ratio of 1:3 and boiled for 5 min. Samples were loaded onto a 384-well plate (Genetix, UK), where each sample was serially diluted five times in 1× SDS buffer. Samples were robotically spotted onto nitrocellulose-coated glass slides (FAST Slides, Whatman, Schleicher and Schuell) using a microarrayer (MicroGridII). Slides were incubated overnight in blocking solution [1 g I-block (Tropix, Bedford, MA, USA), 0.1 % Tween-20 in 500 ml PBS] at 4 °C with constant rocking. After washing three times 5 min each, the slides were incubated with primary antibodies. Table 1 lists the used primary antibodies and their sources/suppliers in addition to β-actin antibody as a control of protein loading, diluted 1:500 in the same antibody diluent overnight at 4 °C with shaking. All the used antibodies were diluted as 1:50.
Following washing, the slides were incubated with diluted infrared secondary (800 CW anti-rabbit) antibody for 30 min at room temperature in dark with shaking. Slide were washed as before, dried by centrifugation at 500× for 5 min and scanned with a Licor Odyssey scanner (LI-COR, Biosciences) at 21 μm resolution at 800 nm (green). The resultant TIFF images were processed with Axon Genepix Pro-6 Microarray Image Analysis software (Molecular Services Inc.) to obtain fluorescence data for each feature. Protein signals were finally determined with background subtraction and normalization to the internal housekeeping targets using RPP analyzer, a module within the R statistical language on the CRAN (http://cran.rproject.org/).
Statistical analysis
Statistical analysis was performed using SPSS 16.0 statistical software (SPPS Inc., Chicago, IL, USA). Kaplan–Meier plots were used to visualise the survival distribution of the resulting clusters, with differences in survival estimated using Log-rank tests. Cox proportional hazards model was fitted to test confounders and statistical independence and significance of these clusters as predictors of BCSS and DMFS.
Results
To address the contribution of E-cadherin loss (E-cad−) and N-cadherin gain (N-cad+) with relevance to specific cancer classes and their impact on patients’ outcome, tumours were defined based on their expression of both proteins. The H scores of both markers did not follow normal Distribution. Therefore, E-cadherin and N-cadherin expressions were dichotomised at their median H-score expressions (=100, where; E-cad or N-cad <100 was considered negative and ≥100 was positive expression). This resulted in four combinatorial phenotypic groups (Table 2) as follows;
- Group 1::
-
E-cad+/N-cad−,
- Group 2::
-
E-cad−/N-cad−,
- Group 3::
-
E-cad+/N-cad+,
- Group 4::
-
E-cad−/N-cad+.
Associations were noticed between these phenotypes and tumour grade (p < 0.001), tumour size and the Nottingham prognostic index (NPI) (p < 0.001) (Table 3). The association with the histological tumour type was statistically significant (p = 0.001); up to 39 % of tubular mixed carcinomas and special type tumours of excellent prognosis were E-cad+/N-cad−, denoting intact E-cad with no gain of N-cad. However, up to 43 % of medullary carcinomas were E-cad−/N-cad+, denoting E-cad− and N-cad up-regulation.
E-cad/N-cad combinatorial groups and BC Molecular subtypes
A significant association was observed between E-cad/N-cad combinatorial groups and breast cancer molecular classes (χ2 = 41.56, p < 0.001) defined as: luminal (ER+ and/or PR+/HER2−), HER2+ (HER2 positive), Triple negative (TN; ER−, PR−, HER2−) including basal (BLBC) (TN and positive for CK5/6, and/or EGFR and/or CK14) and non-basal (TN-non-basal) (negative for all five markers) [28, 35]. In this respect more than one-third (35.4 %) of the luminal class were E-cad+/N-cad−, compared to up to one-fourth (24.4 % and 24.2 %, respectively) of cases in both E-cad−/N-cad− and E-cad+/N-cad+, of the HER2+ and TN subtypes, and one-fifth (20.7 %) in the E-cad−/N-cad+ group in the TN-basal subtype (Table 3).
IHC Expression of cadherin switch phenotypic groups and expression patterns of EMT potential triggers
TGFβ1 was found to be differentially expressed among the different E-cad/N-cad combinatorial groups (ANOVA, F = p < 0.001). It was found to be maximally expressed in the E-cad−/N-cad− and E-cad+/N-cad+ groups, while least expressed in the E-cad+/N-cad−. For pAkt-S473, statistically significant differences were observed between E-cad/N-cad combinatorial groups, with maximal expression in the E-cad−/N-cad− and E-cad+/N-cad+ groups while least expressed in the E-cad+/N-cad−. Similarly, statistically significant differences were observed between E-cad/N-cad combinatorial groups regarding the expression of PIK3CA, TWIST2 and CTEN with the differential expression patterns similar to those of TGFβ1 and pAkt-S473.
E-cad/N-cad combinatorial groups and patients outcome
Significant association is identified between these combinatorial phenotypes and patients outcome in terms of BCSS (LR = 9.09, p = 0.028) and DMFS (LR = 8.82, p = 0.032). Interestingly, BCSS of patients with E-cad+/N-cad− phenotype, signifying intact E-cad without N-cad+, was not different from E-cad−/N-cad− phenotype (up to 70 % survival rate) (p > 0.05). Similarly, survival of E-cad−/N-cad+, representing E-cad− and N-cad+ was not different from E-cad+/N-cad+ phenotype representing intact E-cad with N-cad+; (58 % survival rate). Similar patterns of metastasis free survival overlap between these phenotypes were noticed after 10 years of follow-up (Fig. 1).
Results of RPPA
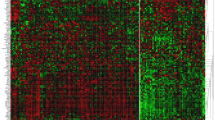
Figure 2 displays scanned images of RPPA slides of the studied proteins. The association between protein expression levels of a specific protein (PIK3CA) and its downstream effecter (pAkt-s473) showed significant direct association (Spearman’s R = 0.798, p < 0.001). Consistent with IHC, RPPA showed variable mean expression levels of these proteins amongst different molecular classes. E-cad expression level was highly expressed in the luminal class (mean = 394.11 pixel) with HER2+ class expressed the lowest level, contrary to N-cadherin that was highly up-regulated in HER2+ while minimally expressed in luminal breast cancer (mean = 2,318.22 pixel and 1,412.26 pixel, respectively). In addition, pAkt-S473 was maximally expressed in luminal breast cancers (mean = 74.86 pixel) while minimally expressed in TN-basal class (36.15 pixel) with HER2+ expressing intermediate values (mean = 47.72 pixel). PIK3CA was maximally expressed in HER2+ (mean = 26.75 pixel). However, TGFβ1 was maximally expressed in TN-non-B followed by the HER2+ (mean = 4,119.77 and 4,015.29 pixel, respectively); Fig. 3.
RPPA analysis for EMT markers/triggers’ proteins in breast cancer samples utilising proteins extracted from macro-dissected FFPE tissues. Proteins were extracted from macro-dissected FFPE samples (n = 52; 49 invasive and 3 in situ BC) and immobilized onto nitrocellulose slides at distint positions. Each sample was printed in duplicate. A positive control lysate (SK-HEp1 liver adenocarcinoma cell line stimulated with external stimuli (LPS, TNF α, IL-2 and IL-8) was printed onto the array for monitoring immunostaining performance. Also negative control (dilution duffer only) was included in the array
Bar charts representing expression levels of EMT markers/triggers in different breast cancer molecular classes. Y axis represents protein expression level (pixel). a Expression levels of E-cad and pAkt-S473 in different molecular classes. b Expression levels of PIK3CA in different molecular classes. c Expression levels of N-cad and TGFB1 in different molecular classes
Discussion
The vast majority of breast cancer patients die from distant metastatic spread rather than from their primary tumours. Therefore, it is crucial to study the molecular mechanisms underlying metastasis to help developing specific therapeutic targets to halt the metastatic seeding of cancer. One of the mechanisms that may enhance the cancer cells capacity to disseminate is EMT. Through this mechanism, it is believed that the migratory capacity is acquired by the transition of malignant epithelial cells into a mesenchymal-like state, thus enhancing their invasive capabilities [36]. Loss of epithelial characteristics coupled with gain of mesenchymal markers has been reported to feature EMT in different cultured cancer cell lines [5]. These changes have been reported to be initiated by deregulation of diverse molecular pathways. However, these phenotypic changes and their triggering pathways are rarely fully observed in vivo. As invasive lobular carcinoma is typically characterised by loss of E-cadherin expression [37], an anticipated feature of EMT, a series of early invasive non-lobular breast cancer cases (n = 1,035) with long-term clinical follow-up has been chosen to study EMT using a large panel of tissue markers (n = 19) using IHC and RPPA.
Combinatorial phenotypic E-cadherin and N-cadherin groups were significantly associated with tumour grade, tumour size, NPI and histological tumour type. In this respect, larger proportions of tumours of high grade, large tumour size, and poor NPI were of the E-cad−/N-cad+ phenotype, denoting E-cad− and N-cad+. The reverse was true where more proportions of tumours with lower grade, smaller size and better NPI were of the E-cad+/N-cad−, indicating intact E-cad expression without gain of the N-cad. This pattern of association, where E-cad− and N-cad+ were associated with poor prognostic attributes of the tumour. Moreover, regarding the histological tumour types, up to two-fifths of tubular mixed carcinomas and special type tumours of excellent prognosis retained E-cad expression yet deemed negative for N-cad expression. Conversely, similar proportions of medullary carcinomas showed E-cad− with N-cadherin expression. In addition, closely equal proportions (~30 %) of the same histological types were of double negative (E-cad−/N-cad−) phenotype. Therefore, up-regulation of N-cad concomitantly with E-cad− appears to be linked with tumour differentiation, with medullary carcinoma and tubular carcinoma representing two differentiation extremes [38], showing significantly different proportions of cadherin switch phenotypes. Accordingly, these findings point to the privileged occurrence of cadherin switching in medullary carcinoma, a high grade type of breast cancer characterised by a pushing border of invasion and typically display basal-like biological features [39].
A significant association was observed between E-cad/N-cad combinatorial groups and breast cancer molecular classes. In this respect, more than one-third of the luminal class was of the E-cad+/N-cad− phenotype compared to up to one-fourth of cases in both of the HER2+ and TN subtypes. Contrasting this, the HER2+, the TN, and TN-non B classes; in order, displayed higher proportions of the E-cad−/N-cad+ phenotype. Therefore, intact E-cad, without N-cad+, was more observed the luminal BC in contrast to other BC classes which displayed higher proportions with E-cad− and N-cad+. Interestingly, findings of RPPA study in a smaller subset of cases, although did not reach statistical significance, showed differential expression values of E-cad and N-cad amongst different BC molecular subtypes. Their expression patterns recapitulated the findings observed using IHC in the larger BC series. E-cadherin expression levels were reciprocally associated with N-cadherin in both luminal and HER2+ breast cancer classes; where down-regulation of the former in HER2+ was coupled with up-regulation of the latter. Also, luminal classes displayed the highest levels of E-cadherin and lowest level of N-cadherin. Therefore, the RPPA technique which can monitor subtle changes in protein expression on quantitative basis revealed results denoting the privileged occurrence of cadherin switch in certain BC subtypes compared to others. These findings validate quantitatively the findings observed and discussed within the context of EMT markers/triggers [40]. In addition, these findings might explain, at least in part, the adverse clinical outcome and the inherent tendency of the HER2+, TN and TN-nonB classes for progression.
Regarding EMT proposed triggers, immunohistochemical expression of TGFβ1 was found to be significantly expressed within cadherin switch combinatorial groups with maximal expression in phenotypes with N-cad+ irrespective of E-cad status. Moreover, IHC expression of pAkt-S473, PIK3CA, TWIST2 and CTEN were significantly expressed between these combinatorial groups with patterns similar to those of TGFβ1. Therefore, translational changes of cadherin switch appear to occur synergistically with activation of major molecular pathways; TGFβ1, Twist2, PIK3CA and pAkt-S473; pathways and transcription factors reported to drive EMT program and probably mainly through up-regulation of N-cad according to the results of the current study [14, 16, 17, 30].
Moreover, cadherin switch combinatorial phenotypes displayed significantly different BCSS and MFS, with E-cad+/N-cad− showed the longest and E-cad−/N-cad+ showed the shortest survival. Interestingly, survival of E-cad+/N-cad− phenotype was not different from E-cad−/N-cad− phenotype. Similarly, survival of E-cad−/N-cad+ was not different from E-cad+/N-cad+ phenotype. In other terms, E-cad− appeared not to be the most crucial event required for tumour progression and N-cadherin has a dominant effect over the tumour suppressive E-cadherin. In-vitro studies reported the persistence of proinvasive actions of N-cadherin even in the presence of E-cadherin [19].
These integrative molecular changes challenge some prevailing views that propose repression of E-cadherin [41], which is moderately retained in luminal, HER2+ and TN-Basal BC, or deregulation of a single molecular pathway to be sufficient to explain higher tendency of certain cancer types to disseminate than the other [17]. The findings of this study are in line with our previous report which showed that N-cadherin expression was the most influential amongst other cadherins in determining the EMT phenotype [40].
Findings of this study illustrate, at translational level, the occurrence of characteristic molecular changes indicative of EMT-like profile that could elucidate heterogeneity and varied clinical behaviour of breast cancer, therefore, could help developing targeted therapies against EMT-associated pathways. A subset of hormone receptor positive luminal breast cancer preferentially displayed EMT markers/triggers that could explain the indigenous tendency of this subset for progression. Therefore, not only proliferation and level of hormonal receptor expression but also an EMT-like program is an additional indicator of heterogeneity within luminal cancers. Translational molecular changes indicative of an EMT program, at least in the studied series of clinical breast cancer samples, appear to result from integration of a diverse array of molecular pathways including TGFβ1, PIK3/Akt, Twist2 and CTEN. These appear to share variably in EMT execution in different molecular classes. RPPA is a useful method for quantitatively comparing protein expression levels in FFPE breast cancer tissue with relevance to different breast cancer classes and distinct progression phases.
References
Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7(2):131–142
Moustakas A, Heldin CH (2007) Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci 98(10):1512–1520
Thiery JP (2002) Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2(6):442–454
Iwatsuki M et al (2010) Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci 101(2):293–299
Cardiff R (2010) The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia 15(2):225–233
Tomaskovic-Crook E, Thompson EW, Thiery JP (2009) Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res 11(6):213
Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9(4):265–273
Wu Y, Zhou BP (2008) New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 40(7):643–650
Xue C et al (2003) The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res 63(12):3386–3394
Giampieri S et al (2009) Localized and reversible TGF[beta] signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol 11(11):1287–1296
Tarin D (2005) The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res 65(14):5996–6001
Wicki A et al (2006) Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell 9(4):261–272
Tse JC, Kalluri R (2007) Mechanisms of metastasis: epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J Cell Biochem 101(4):816–829
Wendt MK, Smith JA, Schiemann WP (2010) Transforming growth factor-beta-induced epithelial-mesenchymal transition facilitates epidermal growth factor-dependent breast cancer progression. Oncogene 29(49):6485–6498
Yang J et al (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117(7):927–939
Lamouille S, Derynck R (2011) Emergence of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin axis in transforming growth factor-beta-induced epithelial-mesenchymal transition. Cells Tissues Organs 193(1–2):8–22
Albasri A et al (2009) C-terminal Tensin-like (CTEN) is an oncogene which alters cell motility possibly through repression of E-cadherin in colorectal cancer. J Pathol 218(1):57–65
Peinado H, Portillo F, Cano A (2004) Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol 48(5–6):365–375
Hazan RB et al (2004) Cadherin switch in tumor progression. Ann N Y Acad Sci 1014:155–163
Ahmed N, Thompson EW, Quinn MA (2007) Epithelial-mesenchymal interconversions in normal ovarian surface epithelium and ovarian carcinomas: an exception to the norm. J Cell Physiol 213(3):581–588
Josson S et al (2010) Tumor-stromal interactions influence radiation sensitivity in epithelial-versus mesenchymal-like prostate cancer cells. J Oncol. doi:10.1155/2010/232831
Lin K et al (2010) The role of B-RAF mutations in melanoma and the induction of EMT via dysregulation of the NF-kappaB/Snail/RKIP/PTEN circuit. Genes Cancer 1(5):409–420
Sarrio D et al (2008) Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 68(4):989–997
Spurrier B, Ramalingam S, Nishizuka S (2008) Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc 3(11):1796–1808
Speer R et al (2007) Development of reverse phase protein microarrays for clinical applications and patient-tailored therapy. Cancer Genomics Proteomics 4(3):157–164
Brase JC et al (2010) Increasing the sensitivity of reverse phase protein arrays by antibody-mediated signal amplification. Proteome Sci 8:36
Abd El-Rehim DM et al (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116(3):340–350
Rakha EA et al (2009) Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15(7):2302–2310
Aleskandarany MA et al (2010) Growth fraction as a predictor of response to chemotherapy in node-negative breast cancer. Int J Cancer 126(7):1761–1769
Sasaki K et al (2009) Significance of twist expression and its association with E-cadherin in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 28:158
Bryan RT, Tselepis C (2010) Cadherin switching and bladder cancer. J Urol 184(2):423–431
Yilmaz M, Christofori G (2010) Mechanisms of motility in metastasizing cells. Mol Cancer Res 8(5):629–642
McCarty KS Jr et al (1985) Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109(8):716–721
Nirmalan NJ et al (2009) Development and validation of a novel protein extraction methodology for quantitation of protein expression in formalin-fixed paraffin-embedded tissues using western blotting. J Pathol 217(4):497–506
Cheang MC et al (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14(5):1368–1376
Nguyen DX, Massague J (2007) Genetic determinants of cancer metastasis. Nat Rev Genet 8(5):341–352
Yoder BJ, Wilkinson EJ, Massoll NA (2007) Molecular and morphologic distinctions between infiltrating ductal and lobular carcinoma of the breast. Breast J 13(2):172–179
Ellis IO et al (2005) Pathology reporting of breast disease : a joint document incorporating the third edition of the NHS Breast Screening Programme’s Guidelines for pathology reporting in breast cancer screening and the second edition of the Royal College of Pathologists’ minimum dataset for breast cancer histopathology. In: NHS Cancer Screening Programmes, Royal College of Pathologists, London
Marginean F et al (2010) Histological features of medullary carcinoma and prognosis in triple-negative basal-like carcinomas of the breast. Mod Pathol 23(10):1357–1363
Aleskandarany M et al (2010) Epithelial mesenchymal transition in invasive breast carcinoma: molecular pathways and relation to molecular subtypes. J Pathol 222(S1):S1–S51
Mahler-Araujo B et al (2008) Reduction of E-cadherin expression is associated with non-lobular breast carcinomas of basal-like and triple negative phenotype. J Clin Pathol 61(5):615–620
Acknowledgments
Ola H Negm and M. AH Ahmed are funded by the Egyptian Ministry of High Education.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aleskandarany, M.A., Negm, O.H., Green, A.R. et al. Epithelial mesenchymal transition in early invasive breast cancer: an immunohistochemical and reverse phase protein array study. Breast Cancer Res Treat 145, 339–348 (2014). https://doi.org/10.1007/s10549-014-2927-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-2927-5