Abstract
The renin–angiotensin system (RAS) has been considered to be implicated in the development of breast cancer. However, the results are inconsistent. In this study, we conducted a meta-analysis to assess the association between four polymorphisms, including angiotensin I-converting enzyme (ACE) I/D and A240T, angiotensin II type 1 receptor (AGTR1) A1166C and angiotensinogen (AGT) M235T polymorphisms, and breast cancer risk. Published literature from PubMed, ISI web of science, and Embase databases were retrieved. All studies evaluating the association between ACE I/D, ACE A240T, AGTR1 A1166C, or AGT M235T polymorphism and breast cancer risk were included. Pooled odds ratio (OR) with 95% confidence interval (CI) was calculated using fixed- or random-effects model. Ten studies (1,650 cases and 9,283 controls) on ACE I/D polymorphism, six studies (1,316 cases and 2,632 controls) on ACE A240T polymorphism, three studies (235 cases and 601 controls) on AGTR1 A1166C polymorphism, and two studies (273 cases and 3,547 controls) on AGT M235T polymorphism were included. Overall, the meta-analysis showed no significant association between I/D or A240T polymorphism and breast cancer risk in either genetic model. Further subgroup analysis by ethnicity also revealed non-significant association in Caucasian or Asian populations except for Africans (the statistically significant association for ACE I/D or A240T polymorphism in Africans derived from only one study). A marginally significant association was observed for AGTR1 A1166C polymorphism in Caucasians (CC vs. AA: OR = 0.31, 95% CI 0.10–0.99). In addition, there was a significant association between AGT M235T polymorphism and breast cancer risk in Caucasians (OR = 1.45, 95% CI 1.12–1.88). The present meta-analysis suggested that ACE I/D and A240T polymorphisms might not be a good predictor of breast cancer risk, while AGTR1 A1166C and AGT M235T polymorphisms might be implicated in the pathogenesis of breast cancer. Given the limited sample size, the findings warrant further investigation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common cancer among women worldwide, which accounts for 16% of all female cancers [1]. Breast cancer has led to serious mortality, and is one of the main causes of global health burden. Although environmental factors, such as reproductive (e.g., age at first birth and breastfeeding), behavioral (e.g., hormone-replacement therapy and alcohol consumption), and anthropometric risk factors (e.g., body mass index), could contribute to the increased risk of breast cancer, and genetic factors are also implicated in the pathogenesis of the disease [2, 3]. Up to now, a great number of genetic variants have been identified to be potentially associated with breast cancer risk [4, 5].
The renin–angiotensin system (RAS) is a hormonal signaling mechanism, which is implicated in the regulation of blood pressure and cardiovascular homeostasis. Angiotensin II (Ang II), the main component of the RAS, is converted from angiotensin I (Ang I) via angiotensin I-converting enzyme (ACE). Ang II exerts its physiological effects by binding to two pharmacologically distinct receptors, namely, Ang II type 1 receptor (AGTR1) and Ang II type 2 receptor(AGTR2) [6]. Moreover, AGTR1 is the predominantly subtype to stimulate actions of Ang II on angiogenesis, cell growth, and cell proliferation in tissues, suggesting that the RAS might be involved in carcinogenesis [7].
The ACE gene, located on chromosome 17q23, contains many polymorphisms. The 287-bp Alu insertion/deletion (I/D) polymorphism in intron 16 and the A240T polymorphism in the 5′-flanking region (two polymorphisms are in tight linkage disequilibrium), are the most studied polymorphisms and have been related to ACE levels [8]. Experimental studies showed that Ang II exerted pro-mitotic, pro-proliferative and angiogenic effects [9], and ACE inhibitor could lower the risk of breast cancer in women, although the results have been inconsistent [10]. The angiotensinogen (AGT) gene (located on 1q42-43) includes one polymorphism M235T which results from a T/C transition in exon 2. AGT has two opposite properties, which could either benefit women through inhibiting cell proliferation or increase breast cancer risk by raising Ang II level which promotes angiogenic activity [11, 12]. The A1166C polymorphism with A/C transversion at position 1166 in the 3′untranslated region of AGTR1 gene (located on 3q23) has been extensively studied in various diseases, especially for blood pressure [13].
So far, several studies have explored the association between the polymorphisms of RAS genes and breast cancer risk; however, the conclusions are inconsistent [14–25]. Taking ACE I/D polymorphism, for an example, several studies reported that D allele was positively [15, 20, 21, 23] or reversely [16] associated with breast cancer risk, while others showed no significant association [24]. The discrepancies may be due to many reasons, such as insufficient statistical power, recruitment procedures of the study population, and differences in the genetic and environmental backgrounds. Meta-analysis is a useful method to overcome the disadvantages of individual studies by increasing the statistical power. In this study, we performed a meta-analysis to assess the association between polymorphisms of RAS genes and breast cancer risk.
Materials and methods
Literature and search strategy
We searched the literature databases including PubMed (1950–2010), ISI web of science (1975–2010), and Embase (1966–2010).
The search strategy to identify all possible studies involved using combinations of the following key words: (“renin–angiotensin system” or “RAS” or “angiotensin-converting enzyme” or “ACE” or “Angiotensin II type 1 receptor” or “AGTR1” or “angiotensinogen” or “AGT”) and (“polymorphism” or “variant”) and (“breast cancer”). The reference lists of reviews and retrieved articles were hand-searched. Supplementary data were searched for missing data points. All searches were limited to studies published in English. If more than one article were published using the same case series, only the study with largest sample size was selected. The literature search was updated on December 10, 2010.
Inclusion criteria and data extraction
The studies included in the meta-analysis must meet all the following inclusion criteria: (1) evaluating the association between ACE I/D, ACE A240T, AGTR1 A1166C or AGT M235T polymorphism and the risk of breast cancer; (2) case–control or cohort design; and (3) sufficient data for calculation of odds ratio (OR) with confidence interval (CI). The following information was extracted from each study: (1) name of the first author; (2) year of publication; (3) country of origin; (4) ethnicity; (5) source of control subjects; (6) numbers of cases and controls; and (7) numbers of genotypes for four polymorphisms in cases and controls. Two authors independently assessed the articles for compliance with the inclusion/exclusion criteria, resolved disagreements, and reached a consistent decision.
Statistical analysis
The association between four polymorphisms of RAS genes and the risk of breast cancer was estimated by calculating pooled OR and 95% CI. The significance of the pooled OR was determined by Z test (P < 0.05 was considered statistically significant). The Q test was performed to evaluate whether the variation was due to heterogeneity or by chance. A random- (DerSimonian–Laird method [26]) or fixed-(Mantel–Haenszel method [27]) effects model was used to calculate pooled effect estimates in the presence (P ≤0.10) or absence (P > 0.10) of heterogeneity, respectively. Subgroup analyses were performed by ethnicity. Sensitivity analysis was performed to evaluate the stability of the results by removing the studies not in Hardy–Weinberg equilibrium. Publication bias was assessed by Egger’s test [28] (P < 0.05 was considered statistically significant). Data analysis was performed using STATA version 11 (StataCorp LP, College Station, Texas, USA).
Results
Characteristics of studies
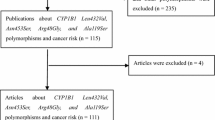
The literature search identified a total of 77 potential relevant articles (Fig. 1). Of these, 65 were excluded because of obvious irrelevance by reading their titles and abstracts. Thus, 12 articles met the inclusion criteria. However, the study by Koh et al. [14] was excluded because of examining the associations of three AGR1 polymorphisms (namely A168G, C535T, and T825A) rather than A1166C polymorphism. The three pairs of articles [15, 17–21] contained the overlapping data, and then the article by Yuan et al. [17] was excluded because of lacking data for calculation of OR with 95% CI; the other two articles by González-Zuloeta Ladd et al. [18] and Yaren et al. [19] were excluded as reporting relatively small sample size. In addition, since the article by Haiman et al. [16] included four case–control studies with different ethnic/racial groups, they were regarded as separate studies in the following meta-analysis. Thus, ten studies [15, 16, 20–24] on ACE I/D polymorphism, six studies [15, 16, 23] on ACE A240T polymorphism, three studies [22–24] on AGTR1 A1166C polymorphism, and two studies [23, 25] on AGT M235T polymorphism were included in the final meta-analyses. Of these studies, seven were on Caucasians [16, 20–24], two were on Asians [15, 16], and one was on Africans [16] for I/D polymorphism; three were on Caucasians [16, 23], two were on Asians [15, 16], and one was on Africans [16] for A240T polymorphism; all three studies [22–24] were on Caucasians for AGTR1 A1166C polymorphism; all two studies [23, 25] were on Caucasians for AGT M235T polymorphism. Genotype distributions in the controls of all studies were in HWE except for two studies [20, 22] for I/D polymorphism, one study [15] for A240T polymorphism and one study [22] for AGTR1 A1166C polymorphism. The characteristics of the included studies are listed in Table 1.
Quantitative data synthesis
For ACE I/D polymorphism, eleven studies consisted of 1,650 cases and 9,283 controls were identified. Overall, the results showed no significant association between I/D polymorphism and breast cancer risk (for DD vs. II: OR = 1.22, 95% CI 0.81–1.84; for ID vs. II: OR = 0.85, 95% CI 0.69–1.05; for dominant model: OR = 0.97, 95% CI 0.76–1.22; for recessive model: OR = 1.32, 95% CI 0.86–2.04) (Table 2). In the subgroup analysis by ethnicity, no significant association was observed in all genetic models except for the association in Africans (the statistically significant association for ACE I/D polymorphism in Africans derived from only one study) (Table 2). Sensitivity analysis was performed after excluding the two studies by Yaren et al. [20] and Alves Corrêa et al. [22] deviated from HWE, and the results were not materially altered for I/D polymorphism in either genetic model (Table 2).
For ACE A240T polymorphism, six studies comprised 1,316 cases and 2,632 controls were identified. Overall, the results showed no significant association between A240T polymorphism and breast cancer risk (for co-dominant model: TT vs. AA: OR = 1.06, 95% CI 0.73–1.55, AT vs. AA: OR = 1.03, 95% CI 0.89–1.20; for dominant model: OR = 1.04, 95% CI 0.90–1.19; for recessive model: OR = 1.05, 95% CI 0.73–1.52) (Table 3). In the subgroup analysis by ethnicity, no significant association was observed in all genetic models except for the association in Africans (the statistically significant association for ACE A240T polymorphism in Africans derived from only one study) (Table 3). Sensitivity analysis excluding the study by Koh et al. [15] not in HWE further confirmed the null association (Table 3).
For AGTR1 A1166C polymorphism, three studies comprised 235 cases and 601 controls were identified. Significant association between A1166C polymorphism and breast cancer risk was observed for AC versus AA and dominant model. However, after excluding the study by Alves Corrêa et al. [22] not in HWE, the association disappeared for AC versus AA and dominant model; while a marginally significant association was observed for CC versus AA (OR = 0.31, 95% CI 0.10–0.99) (Table 4).
For AGT M235T polymorphism, two studies comprised 273 cases and 3,547 controls were identified. As the study by González-Zuloeta Ladd [25] did not present the exact frequencies of genotypes in both cases and controls, and it just presented OR with 95% CI for MM versus MT + TT, which was 1.4 (1.1–1.9), thus, we calculated the summary OR with 95% CI under recessive model. There was significant association between M235T polymorphism and breast cancer risk (OR = 1.45, 95% CI 1.12–1.88).
Publication bias
Egger’s test was performed to assess potential publication bias for ACE I/D polymorphism. No publication bias was detected among the included studies (P = 0.07 in homozygous co-dominant genetic model; P = 0.66 in heterozygous co-dominant genetic model; P = 0.09 in dominant genetic model; and P = 0.22 in recessive genetic model). We did not assess the publication bias for ACE A240T, AGTR1 A1166C, or AGT M235T polymorphism based on the knowledge of Cochrane Handbook for Systematic Reviews of Interventions (www.cochranehandbook.org) which states that the test for publication bias yields unreliable results when less than 10 studies are included in a meta-analysis.
Discussion
To the best of our knowledge, this study represents the first meta-analysis of the association between polymorphisms of RAS genes (ACE I/D, ACE A240T, AGTR1 A1166C, and AGT M235T polymorphisms) and breast cancer risk. The findings suggested that ACE I/D and A240T polymorphisms were not likely to be implicated in the development of breast cancer among Caucasians and Asians, except for Africans; while AGTR1 A1166C and AGT M235T polymorphisms might play a role in breast cancer risk among Caucasians. However, the conclusions should be made with caution because of the limited sample size, especially for the statistically significant association for ACE I/D and A240T polymorphisms in Africans.
Many studies supported that the RAS had an important role in the regulation of cell proliferation, angiogenesis, and inflammation [29], suggesting that RAS genes might be implicated in the carcinogenesis [7]. Up to now, ACE I/D is the exclusively studied polymorphism which might be related to breast cancer risk, while with conflicting results. Koh et al. [15] first reported that women with I allele had decreased risk of breast cancer in Chinese (ID + II vs. DD: OR = 0.54, 95% CI 0.29–0.99). Since then, several other studies were published. The study by Yaren et al. [20] showed that compared with II genotype, ID genotype was more commonly observed in breast cancer patients in Turk population(P = 0.03). A prospective study conducted in Netherlands also demonstrated that DD genotype carriers had an increased risk of breast cancer compared with those with II/ID genotypes (hazard ratio = 1.47, 95% CI 1.05–2.04) [21]. More recently, the study by Mendizábal-Ruiz et al. [23] reported D allele was strongly associated with breast cancer risk (ID + DD vs. II: OR = 5.10, 95% CI 1.79–14.52). Contrary to these findings, in a multiethnic cohort study which included African Americans, Japanese, Latinas, and whites, women with the II genotype was found to have a marginally significant increase in breast cancer risk (II vs. DD: OR = 1.30, 95% CI 1.05–1.61) [16]. Another study showed ID genotype carriers were 3.1 times less likely to develop breast cancer than those with II/DD genotypes in Brazilians [22]. However, recently, one study in Iranian population, suggested that I/D polymorphism had no effect on breast cancer risk (P = 0.15) [24]. Our present meta-analysis suggested that ACE I/D polymorphism might not be a strong predictor of breast cancer risk. A recent meta-analysis of the association between ACE I/D polymorphism and cancer risk also showed that there was no statistically significant association between this polymorphism and breast cancer risk in all combined populations (P > 0.05), although a statistically significant association was observed in all postmenopausal women based on only two published studies (P < 0.05) [30].
So far, there are only three studies [22–24] that have evaluated the association between AGTR1 A1166C polymorphisms and breast cancer, but also yielded inconsistent results. Our meta-analysis showed that CC homozygote might be a protective factor of breast cancer development in Caucasians (CC vs. AA: OR = 0.31, 95% CI 0.10–0.99). However, a recent meta-analysis showed that AGTR1 A1166C allele conferred an increased risk of hypertension (OR = 1.14; 95% CI 1.00–1.30). Therefore, further studies are necessary to explore the association with breast cancer risk for this polymorphism. Other polymorphisms in AGTR1 with breast cancer risk have also been investigated. Three polymorphisms (A168G, C535T, and T825A) in the 5′ region were found positively to be associated breast cancer in a Chinese population [14], while C573T polymorphism was not significantly associated with increased breast cancer risk in a Caucasian population [25].
For AGT M235T polymorphism, González-Zuloeta Ladd et al. [25] reported that MM genotype carriers had higher risk of developing breast cancer (MM vs. MT + TT: OR = 1.4, 95% CI 1.1–1.9), while Mendizábal-Ruiz et al. [23] did not found significant association (MM vs. MT + TT: OR = 1.97, 95% CI 0.87–4.42). Further combined results yielded positive associations, suggesting that AGT M235T polymorphism might be implicated in the development of breast cancer.
In addition, there are several studies investigating the association between ACE gene polymorphism and risk of other cancers, such as gastric cancer [31–34], colorectal cancer [21, 35–37], lung cancer [21, 38, 39], and prostate cancer [21, 40–42]. The results have also been inconsistent. Besides breast cancer, the disparate findings for various cancers could be partly explained by the gene–gene/environment interactions. It is well accepted that dietary and other environmental factors (e.g., use of ACE inhibitor and green tea intake) could influence the association between RAS genes and breast carcinogenesis [14, 17]. In addition, difference in linkage disequilibrium between populations might also explain the conflicting associations [16].
The current meta-analysis has some advantages compared to other individual studies; however, it does have some limitations. First, the present meta-analysis was based primarily on unadjusted effect estimates and CIs (since most studies did not provide the adjusted OR and 95% CI controlling for potential confounding factors), so the effect estimates were relatively imprecise. Second, the effect of gene–gene/gene–environment interactions was not addressed in this meta-analysis. Third, the results of subgroup analysis should be interpreted with caution because of limited statistical power. Fourth, the potential publication bias was not assessed for ACE A240T, AGTR1 A1166C, or AGT M235T polymorphism because of limited number of studies. Thus, we can not exclude the possibility of publication bias for these polymorphisms.
In summary, ACE gene I/D and A240T polymorphisms might not be a good predictor of breast cancer risk, while AGTR1 A1166C and AGT M235T polymorphisms might be implicated in the pathogenesis of breast cancer. However, given the limited data, it is not possible to draw conclusions on the exact risk of breast cancer associated with RAS genes, which warrant further investigation.
References
Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55(2):74–108
Travis RC, Reeves GK, Green J, Bull D, Tipper SJ, Baker K, Beral V, Peto R, Bell J, Zelenika D, Lathrop M, Million Women Study Collaborators (2010) Gene–environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet 375(9732):2143–2151
Narod SA (2010) Genes, the environment, and breast cancer. Lancet 375(9732):2123–2124
Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M (2010) Genetic susceptibility to breast cancer. Mol Oncol 4(3):174–191
Eccles D, Tapper W (2010) The influence of common polymorphisms on breast cancer. Cancer Treat Res 155:15–32
Timmermans PB, Chiu AT, Herblin WF, Wong PC, Smith RD (1992) Angiotensin II receptor subtypes. Am J Hypertens 5(6 Pt 1):406–410
Deshayes F, Nahmias C (2005) Angiotensin receptors: a new role in cancer? Trends Endocrinol Metab 16(7):293–299
Villard E, Tiret L, Visvikis S, Rakotovao R, Cambien F, Soubrier F (1996) Identification of new polymorphisms of the angiotensin I-converting enzyme (ACE) gene, and study of their relationship to plasma ACE levels by two-QTL segregation-linkage analysis. Am J Hum Genet 58(6):1268–1278
Sadoshima J, Aoki H, Izumo S (1997) Angiotensin II and serum differentially regulate expression of cyclins, activity of cyclin-dependent kinases, and phosphorylation of retinoblastoma gene product in neonatal cardiac myocytes. Circ Res 80(2):228–241
Lever AF, Hole DJ, Gillis CR, McCallum IR, McInnes GT, MacKinnon PL, Meredith PA, Murray LS, Reid JL, Robertson JW (1998) Do inhibitors of angiotensin-I-converting enzyme protect against risk of cancer? Lancet 352(9123):179–184
Célérier J, Cruz A, Lamandé N, Gasc JM, Corvol P (2002) Angiotensinogen and its cleaved derivatives inhibit angiogenesis. Hypertension 39(2):224–228
Gould AB, Green D (1971) Kinetics of the human renin and human substrate reaction. Cardiovasc Res 5(1):86–89
Niu W, Qi Y (2010) Association of the angiotensin II type I receptor gene +1166 A>C polymorphism with hypertension risk: evidence from a meta-analysis of 16474 subjects. Hypertens Res 33(11):1137–1143
Koh WP, Yuan JM, Van Den Berg D, Lee HP, Yu MC (2005) Polymorphisms in angiotensin II type 1 receptor and angiotensin I-converting enzyme genes and breast cancer risk among Chinese women in Singapore. Carcinogenesis 26(2):459–464
Koh WP, Yuan JM, Sun CL, van den Berg D, Seow A, Lee HP, Yu MC (2003) Angiotensin I-converting enzyme (ACE) gene polymorphism and breast cancer risk among Chinese women in Singapore. Cancer Res 63(3):573–578
Haiman CA, Henderson SO, Bretsky P, Kolonel LN, Henderson BE (2003) Genetic variation in angiotensin I-converting enzyme (ACE) and breast cancer risk: the multiethnic cohort. Cancer Res 63(20):6984–6987
Yuan JM, Koh WP, Sun CL, Lee HP, Yu MC (2005) Green tea intake, ACE gene polymorphism and breast cancer risk among Chinese women in Singapore. Carcinogenesis 26(8):1389–1394
González-Zuloeta Ladd AM, Arias Vásquez A, Sayed-Tabatabaei FA, Coebergh JW, Hofman A, Njajou O, Stricker B, van Duijn C (2005) Angiotensin-converting enzyme gene insertion/deletion polymorphism and breast cancer risk. Cancer Epidemiol Biomarkers Prev 14(9):2143–2146
Yaren A, Turgut S, Kursunluoglu R, Oztop I, Turgut G, Kelten C, Erdem E (2006) Association between the polymorphism of the angiotensin-converting enzyme gene and tumor size of breast cancer in premenopausal patients. Tohoku J Exp Med 210(2):109–116
Yaren A, Turgut S, Kursunluoglu R, Oztop I, Turgut G, Degirmencioglu S, Kelten C, Erdem E (2007) Insertion/deletion polymorphism of the angiotensin I-converting enzyme gene in patients with breast cancer and effects on prognostic factors. J Investig Med 55(5):255–261
van der Knaap R, Siemes C, Coebergh JW, van Duijn CM, Hofman A, Stricker BH (2008) Renin–angiotensin system inhibitors, angiotensin I-converting enzyme gene insertion/deletion polymorphism, and cancer: the Rotterdam Study. Cancer 112(4):748–757
Alves Corrêa SA, Ribeiro de Noronha SM, Nogueira-de-Souza NC, Valleta de Carvalho C, Massad Costa AM, Juvenal Linhares J, Vieira Gomes MT, Guerreiro da Silva ID (2009) Association between the angiotensin-converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms and breast cancer among Brazilian women. J Renin Angiotensin Aldosterone Syst 10(1):51–58
Mendizábal-Ruiz AP, Morales JA, Castro Marti Nez X, Gutierrez Rubio SA, Valdez L, Vásquez-Camacho JG, Sanchez Corona J, Moran Moguel MC (2010) RAS polymorphisms in cancerous and benign breast tissue. J Renin Angiotensin Aldosterone Syst. doi: 10.1177/1470320310383735
Namazi S, Monabati A, Ardeshir-Rouhani-Fard S, Azarpira N (2010) Association of angiotensin I converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms with breast cancer prognostic factors in Iranian population. Mol Carcinog 49(12):1022–1030
González-Zuloeta Ladd AM, Arias Vásquez A, Siemes C, Yazdanpanah M, Coebergh JW, Hofman A, Stricker BH, van Duijn CM (2007) Differential roles of Angiotensinogen and Angiotensin Receptor type 1 polymorphisms in breast cancer risk. Breast Cancer Res Treat 101(3):299–304
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22(4):719–748
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
Ager EI, Neo J, Christophi C (2008) The renin–angiotensin system and malignancy. Carcinogenesis 29(9):1675–1684
Ruiter R, Visser LE, Van Duijn CM, Ch Stricker BH (2011) The ACE insertion/deletion polymorphism and risk of cancer, a review and meta-analysis of the literature. Curr Cancer Drug Targets 11(4):421–430
Röcken C, Lendeckel U, Dierkes J, Westphal S, Carl-McGrath S, Peters B, Krüger S, Malfertheiner P, Roessner A, Ebert MP (2005) The number of lymph node metastases in gastric cancer correlates with the angiotensin I-converting enzyme gene insertion/deletion polymorphism. Clin Cancer Res 11(7):2526–2530
Ebert MP, Lendeckel U, Westphal S, Dierkes J, Glas J, Folwaczny C, Roessner A, Stolte M, Malfertheiner P, Röcken C (2005) The angiotensin I-converting enzyme gene insertion/deletion polymorphism is linked to early gastric cancer. Cancer Epidemiol Biomarkers Prev 14(12):2987–2989
Goto Y, Ando T, Nishio K, Ishida Y, Kawai S, Goto H, Hamajima N (2005) The ACE gene polymorphism is associated with the incidence of gastric cancer among H. pylori seropositive subjects with atrophic gastritis. Asian Pac J Cancer Prev 6(4):464–467
Sugimoto M, Furuta T, Shirai N, Ikuma M, Sugimura H, Hishida A (2006) Influences of chymase and angiotensin I-converting enzyme gene polymorphisms on gastric cancer risks in Japan. Cancer Epidemiol Biomarkers Prev 15(10):1929–1934
Röcken C, Neumann K, Carl-McGrath S, Lage H, Ebert MP, Dierkes J, Jacobi CA, Kalmuk S, Neuhaus P, Neumann U (2007) The gene polymorphism of the angiotensin I-converting enzyme correlates with tumor size and patient survival in colorectal cancer patients. Neoplasia 9(9):716–722
Nikiteas N, Tsigris C, Chatzitheofylaktou A, Yannopoulos A (2007) No association with risk for colorectal cancer of the insertion/deletion polymorphism which affects levels of angiotensin-converting enzyme. In Vivo 21(6):1065–1068
Toma M, Cimponeriu D, Apostol P, Stavarachi M, Cojocaru M, Belusică L, Crăciun AM, Radu I, Gavrilă L (2009) Lack of association between ACE ID polymorphism and colorectal cancer in Romanian patients. Chirurgia (Bucur) 104(5):553–556
Cheon KT, Choi KH, Lee HB, Park SK, Rhee YK, Lee YC (2000) Gene polymorphisms of endothelial nitric oxide synthase and angiotensin-converting enzyme in patients with lung cancer. Lung 178(6):351–360
Nacak M, Nacak I, Sanli M, Ozkur M, Pektaş M, Aynacioğlu AS (2010) Association of angiotensin converting enzyme gene insertion/deletion polymorphism with lung cancer in Turkey. Cancer Genet Cytogenet 198(1):22–26
Medeiros R, Vasconcelos A, Costa S, Pinto D, Lobo F, Morais A, Oliveira J, Lopes C (2004) Linkage of angiotensin I-converting enzyme gene insertion/deletion polymorphism to the progression of human prostate cancer. J Pathol 202(3):330–335
Yigit B, Bozkurt N, Narter F, Yilmaz H, Yucebas E, Isbir T (2007) Effects of ACE I/D polymorphism on prostate cancer risk, tumor grade and metastatis. Anticancer Res 27(2):933–936
Sierra Díaz E, Sánchez Corona J, Rosales Gómez RC, Gutierrez Rubio SA, Vázquez Camacho JG, Solano Moreno H, Morán Moguel MC (2009) Angiotensin-converting enzyme insertion/deletion and angiotensin type 1 receptor A1166C polymorphisms as genetic risk factors in benign prostatic hyperplasia and prostate cancer. J Renin Angiotensin Aldosterone Syst 10(4):241–246
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xi, B., Zeng, T., Liu, L. et al. Association between polymorphisms of the renin–angiotensin system genes and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 130, 561–568 (2011). https://doi.org/10.1007/s10549-011-1602-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1602-3