Abstract
In this article, an attempt for efficient screening of circulating tumor cells (CTCs) with excellent operability on microfluidic chips was reported. A Parsortix™-like cell separation chip was manufactured in our lab. This chip allowed lateral flow of fluid which increased the flow rate of blood. And, an air valve controlled injection pump was manufactured which allowed eight chips working simultaneously. This greatly facilitated the blood treatment process and saved time. As for the mechanism of screening circulating tumor cells, selective size amplification was utilized. By size amplification of cancer cells, both the hardness and the size of CTCs increased which differentiated them from blood cells. And the modification procedure of beads used for size amplification of cancer cells was optimized. Finally, by integrating the commercialized Parsortix™-like cell separation chip and selective size amplification, a practical method for screening circulating tumor cells was established.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Circulating tumor cells (CTCs) are regarded as escaped tumor cells from tumor site into blood (Cristofanilli et al. 2004; de Bono et al. 2008; Hyun et al. 2016). They carry large amount of information about cancer (Wang et al. 2016; Xu et al. 2017). Understanding them can help us propose efficient therapeutic schedule. However, the main obstacle in front of us is how to capture them as they are extremely rare in blood (Chen et al. 2014). Facing this problem, lots of work has been done to concentrate the CTCs (Patil et al. 2015; Jin et al. 2014; Khoo et al. 2014). For example, by tuning the size of micro-sieves, some large sized cells are concentrated and captured (Lim et al. 2012). By gradient centrifugation, most of the eukaryotic cells including white blood cells (WBCs) and CTCs are enriched (Song et al. 2017; Liao et al. 2017). By erythrocyte lysis, nearly all of the red blood cells (RBCs) are removed (Arya et al. 2013). But, there still exists some problems such as loss of target cells when processing the blood. And the commercial product such as CELLSEARCH® has been applied in some developed countries, but it costs too much (Riethdorf et al. 2007; Liu et al. 2016; Kim et al. 2016).
With the development of soft-lithography, Toner firstly reported a new tool to capture CTCs by using microfluidic chips (Zhao et al. 2013; Nagrath et al. 2007; Stott et al. 2010; Yu et al. 2013). By modifying specific antibody anti-EpCAM onto the channel surface, they successfully captured CTCs. Since then, some methods based on microfluidic chips are developed including positive and negative screening (Li et al. 2015; Kang et al. 2012; Karabacak et al. 2014; Chaudhuri et al. 2016). However, as an emerging technique, there are still some problems to solve. For example, the inner structure of microfluidics is sophisticated (Guan et al. 2013; Ozkumur et al. 2013; Warkiani et al. 2014; Zhang et al. 2016). What is worse is that the operability is very poor and clogging often occurs as the blood is a highly viscous liquid (Chaudhuri et al. 2016). Therefore, it is necessary to develop a practical method for efficient screening of CTCs.
In this work, we integrated the commercialized chips and leading-edge size amplification technology for practical screening of CTCs with excellent operability. In detail, the commercialized Parsortix™-like cell separation chip which was convenient to manufacture was utilized (Hvichia et al. 2016). And selective size amplification (SSA) as an effective method to differentiate cancer cells from white blood cells was also applied (Kim et al. 2012). The Parsortix™-like cell separation chip was manufactured in our lab. This chip permitted lateral flow of blood which increased the fluid cross section. And an air valve controlled sample injection system was applied which allowed eight chips working simultaneously. This largely increased the flow rate of blood in microfluidic chips. As for the SSA method, we compared two modification methods of beads by anchoring antibody onto them. And we gave an optimized size amplification procedure. Finally, a practical method for screening CTCs with excellent operability was given.
2 Experiments
2.1 Materials
Parsortix™-like cell separation chip was manufactured in ultraclean room; streptavidin modified beads (Dynabeads™ M-280, 2.8 μm, 10 mg mL−1, ~6–7 × 108 mL−1) were purchased from Thermo Fisher Scientific Company; biotin anti-human CD326 (EpCAM) antibody was purchased from BioLegend Company; carboxylate-modified polystyrene beads (diameter 2.0–2.9 μm, 2.5% w/v) were purchased from Aladdin Company; MCF-7 cancer cells were obtained from American Type Culture Collection (ATCC; Manassas, VA) and cultured as the given instruction: they were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 IU mL−1 penicillin, and 100 mg mL−1 streptomycin; fetal bovine serum (FBS), phosphate buffer solution (PBS, pH 7.4), hoechst 33342, biotin-conjugated rabbit IgG and FITC-conjugated donkey anti-rabbit IgG were purchased from Sangon Biotech Company.
2.2 Instrument
Fluorescence microscope (Leica AF6000) was used to investigate the captured cancer cells; 3D laser scanning confocal microscope (VK-X Series) was used to measure the outline of the microfluidic chips; air valve controlled injection pump for injecting blood with high flow velocity was manufactured in our lab.
2.3 Chip fabrication
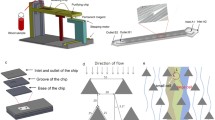
The Parsortix™-like cell separation chip (A microfluidic chip for direct and rapid trapping of white blood cells from whole blood (Chen et al. 2013)) was fabricated according to the reported method with some modification. First, a silicon wafer with 480 μm thickness was used as the substrate, as shown in Fig. 1a. A Cr/Cu seed layer was sputtered on the silicon wafer for the subsequent electroplating, as shown in Fig. 1b. Then silicon wafer was spin coated with positive photoresist (AZ 4903) about 5 μm in thickness and patterned, as shown in Fig. 1c. Cu about 5 μm in thickness was electroformed to fabricate the channel and pillars of trapping structures, as shown in Fig. 1d. After removing the positive photoresist, the wafer was spin coated with negative photoresist (SU-8) about 30 μm in thickness and patterned again to fabricate the main channel. The mother mold with inverse structures was fabricated after removal of the photoresist, as shown in Fig. 1e. Then, a 10:1 ratio of the PDMS monomer and a curing agent (Sylgard 184, Dow Corning) was poured over the mother mold and baked at 70 °C for 2 h, as shown in Fig. 1f. After PDMS was released from the mold and punched through to function as inlets/outlet for sample injection and extraction, PDMS was bonded with a glass slide after oxygen plasma treatment, as shown in Fig. 1g.
Fabrication process of the Parsortix™-like cell separation chip. (a) A Si wafer; (b) sputtering a Cr/Cu seed layer; (c) patterning positive photoresist; (d) electroforming Cu; (e) patterning negative photoresist; (f) transferring the structure to PDMS slices; and (g) punching inlets/outlets and bonding PDMS slices onto a glass slide
2.4 Modification of the Dynabeads
Dynabeads™ M-280 was modified by biotinylated anti-human CD326 (EpCAM) antibody according to the manufacture’s instruction. The modification process was shown below: 20 μL of dynabeads was diluted in 5 mL of PBS containing 0.1% bovine serum albumin (BSA) and 0.02% sodium azide and incubated for about 5 min at 37 °C. And then the dynabeads were separated at magnetic field by a magnet. The above procedure was repeated for 3 times and the dynabeads were dispersed in 5 mL of PBS. 2 μL of biotin modified anti-human CD326 antibody was added into this solution and incubated for about 30 min at 37 °C. After incubation, the modified dynabeads were separated at magnetic field by a magnet. The separated dynabeads were dispersed in 5 mL of PBS solution containing 1 vol% of FBS for 3 min at 37 °C and then separated at magnetic field by a magnet. This procedure was repeated for 3 times. The final anti-human CD326 modified dynalbeads were dispersed in 100 μL of PBS solution and stored for use at 4 °C refrigerator.
2.5 Size amplification of cancer cells
MCF-7 cells were purchased from the American Type Culture Collection (ATCC) and propagated according to the manufacturer’s instructions. The cells were washed by culture medium and PBS, respectively. And then the cells were counted and diluted in 20,000 mL−1. 5 mL of the cell solution was added into 100 μL solution of anti-human CD326 antibody modified dynabeads and incubated with shaking at 37 °C. 15 min later, this mixture was centrifuged at 800 rpm for 10 min to clear away the unreacted beads. Finally, the centrifuged cells were investigated by microscope.
2.6 Cell screening
7.5 mL of whole blood containing MCF-7 cancer cells (10, 100, and 1000 mL−1) was firstly mixed with 100 μL solution of anti-human CD326 modified dynabeads and incubated at 37 °C for about 30 min. And then, this blood sample was diluted by 5 multiples with PBS. Finally, it was pumped into the manufactured microfluidic chip by air valve controlled injection pump. After pumping the blood sample, the captured cancer cells was washed by PBS on chip and then stained by hoechst 33342. And the captured cancer cells under fluorescence microscope were counted and the capture efficiency was calculated.
3 Results and discussion
3.1 Fabrication of Parsortix™-like cell separation chip
Manufacture of Parsortix™-like cell separation chip mold was in super-clean room. The procedure was shown in Fig. 1. This was a double-layered mold as shown in Fig. 2a. This mold contained channel, barrier and pillar. The size of them could be adjusted as needed. In our experiments, the width of both the channel and barrier was set as 200 μm. Increasing the width of the channel would lead to the collapse of the PDMS layer and increasing the width of the barrier would result in the increasing of fluid resistance. There was gap between the substrate and the PDMS layer as shown in Fig. 1g. The distance of the gap corresponded to the height of the barrier in the chip mold. And the distance of this gap could also be adjusted. Fluid could flow in the channel and pass through the gap. By this manner, some small size cells such as red blood cells (RBCs) and partial of the white blood cells (WBCs) could pass through the gap as shown in Fig. 2c. However, CTCs and large size WBCs could not pass. In our experiments, we carefully explored the lower critical distance of the gap above which all of the cells could pass. We found that when the distance of the gap was above 5 μm, all of the cells could pass through the gap by deformation. And when the distance of the gap was lower than 3 μm, all of the cancer cells and most of the WBCs could not pass through the gap. This would cause severe clogging in microfluidic chips. The distance of the gap and the channel here we used were set to 4.1 μm and 30 μm, respectively. The final prepared chip was shown in Fig. 2b. The advantage of this chip was that fluid could flow in lateral directions which largely increased the fluid cross-section and avoided clogging as shown in Movie S1.
Injection pump was self-made in our lab as show in Fig. S1. This was an air valve controlled pump. It could allow eight chips working simultaneously. The injection pump was controlled by our mobile phone which served as a remote controller. And the controlling software was also developed by our lab as shown in Fig. S2. This injection pump provided us a convenient instrument to treat blood with high through-put.
3.2 Problems and solutions of screening CTCs
After obtaining the Parsortix™-like cell separation chip, its performance for screening CTCs was tested. 7.5 mL of whole blood from volunteers was pumped into the chip at the flow rate of 20~200 μL h−1. Before pumping the whole blood into the chip, MCF-7 cancer cells were added into the whole blood by the concentration of 1000 mL−1. The nucleus of cancer cell was stained by hoechst 33342 in advance. During the process, we found that the whole blood was high viscosity liquid which led to the clogging as shown in Fig. 3a-c. And the flow rate could not increase for its high viscosity of blood. Although the CTCs were captured by our chips, the large amount of jammed blood cells still existed. In order to solve the clogging problem, the whole blood was firstly diluted into different multiples by PBS. We found that viscosity of whole blood could decrease dramatically when they were diluted at least 5 multiples. 2–4 multiples dilution was not enough. Unfortunately, the volume of blood also increased after dilution. It needed more time to process them. As a result, hemolysis occurred as shown in Fig. S3. Hemolysis was really a bad result we should avoid. It made CTCs screening nonsense especially considering the following biomedical analysis of captured CTCs. However, the good thing was that the flowing rate of diluted blood could be increased. Although the flowing rate of diluted blood could increase to 1000 μL h−1, the capture efficiency decreased because CTCs could pass through the gap at high flow field as shown in Fig. 3d-f. This was due to cells were deformable under high flow field. The above mentioned problems largely limited the feasibility of screening CTCs. According to previous report (Kim et al. 2012), a size amplification method had been proposed in which the deformability of cancer cell was reduced after combination with beads. By selective size amplification, not only the hardness of cancer cell was enhanced, but also the size of cancer cell increased which differentiated CTCs from WBCs. As an attempt, selective size amplification method was adopted.
3.3 Selective size amplification
In the process of size amplification, modification of beads by specific antibody was necessary. In order to achieve better modification efficiency, two different methods were estimated as shown in Fig. 4a. One method utilized the reaction between carboxyl group on beads and amine group of the antigen/antibody. However, the reaction efficiency was low because no fluorescence could be seen as shown in Fig. 4b. This was due to the activation efficiency of carboxyl group on beads was low. The other method was using specific combination between streptavidin and biotin. It was well known that the binding constant of them was very high. Therefore, the modification efficiency of antibody on the surface was also very high as shown in Fig. 4c. In our experiments, we chose the second method for anchoring anti-EpCAM onto the surface of beads. As for the diameter of beads, we found that when the diameter of beads was larger than 4 μm, it could not pass through the gap. Due to the usage amount of beads was far more than that of cells for efficient binding, it would cause serious clogging when they passed through the gap as shown in Fig. S4. In our experiments, 2.8 μm Dynabeads™ was selected.
Size amplification was an efficient method to differentiate CTCs from WBCs both in size and deformability. And it was also a method to protect CTCs from splitting at high flow field. After modification of beads, combination efficiency of beads with EpCAM expressive MCF-7 cells was estimated as shown in Fig. 5a. Beads could anchor onto the surface of MCF-7 cells easily. After combination, both the size and hardness of cancer cell increased because they could not pass through the gap by deformation even at high flow field as shown in Fig. 5b. But the beads could pass through the gap. And the captured CTCs remained intact.
3.4 Evaluation of capture efficiency
By integrating the Parsortix™-like cell separation chip and selective size amplification method, the capture efficiency of CTCs was estimated. Based on the above results, whole blood containing MCF-7 cancer cells was firstly blended with anti-EpCAM modified beads and incubated at 37 °C for about 30 min. For efficient combination of beads and MCF-7 cancer cells, the amount of beads used was at least 1000 times than cancer cells. And then, this mixture was diluted with PBS by 5 multiples. Finally, this diluted sample was injected into the Parsortix™-like cell separation chip. Compared to the capture efficiency of CTCs without size amplification, it increased dramatically after they were combined with beads as shown in Fig. 6a. The capture efficiency was more than 90% even when the flow rate increased to 3000 μL h−1. And the purity of the captured tumor cells after size amplification increased with increasing the flow rate as shown in Fig. 6b. This was due to the deformability of cancer cells was restrained. But the WBCs could still pass through the gap freely by deformation at high flow rate. For different concentration of CTCs, the capture efficiency decreased with increasing the concentration of CTCs. This was due to the average amount of beads for each of CTC decreased with increasing the concentration of CTCs. Thus, they could also pass through the gap by deformation when cancer cells were anchored by less beads. For one person, it meant that 37.5 mL of diluted blood (equals to 7.5 mL of whole blood) should be processed within 6 h. When the flowing rate was fixed at 3000 μL h−1 and 8 chips were used simultaneously, the whole process time was no more than 2 h, which greatly saved the time.
a Comparison of CTCs capture efficiency before and after combination with beads at different flowing velocity; the blood was diluted by 5 multiples; b Purity of captured CTCs at different flowing rate. For both the capture efficiency and purity experiments, the concentration of CTCs was fixed at 100 mL−1
4 Conclusions
In this work, we presented an efficient method to capture circulating tumor cells by integrating commercialized microfluidic chips and mature selective size amplification method. We also used self-made air valve controlled injection pump to process the blood and software to control the injection pump. It could allow eight chips working simultaneously. The size amplification method was optimized for high efficient modification of anti-EpCAM and combination with cancer cells. In our experiments, the capture efficiency increased after selective size amplification and the purity also increased with increasing the flowing rate. Finally, the reported method to screen CTCs could be finished within 2 h.
References
S.K. Arya, B. Lim, A.R.A. Rahman, Lab Chip 13, 1995–2027 (2013)
P.K. Chaudhuri, M. Ebrahimi Warkiani, T. Jing, Kenry, C.T. Lim, Analyst 141, 504–524 (2016)
J. Chen, D. Chen, T. Yuan, Y. Xie, X. Chen, Biomicrofluidics, 7 (2013)
Y.C. Chen, P. Li, P.H. Huang, Y.L. Xie, J.D. Mai, L. Wang, N.T. Nguyen, T.J. Huang, Lab Chip 14, 626–645 (2014)
M. Cristofanilli, G.T. Budd, M.J. Ellis, A. Stopeck, J. Matera, M.C. Miller, J.M. Reuben, G.V. Doyle, W.J. Allard, L. Terstappen, D.F. Hayes, N. Engl. J. Med. 351, 781–791 (2004)
J.S. de Bono, H.I. Scher, R.B. Montgomery, C. Parker, M.C. Miller, H. Tissing, G.V. Doyle, L.W.W.M. Terstappen, K.J. Pienta, D. Raghavan, Clin. Cancer Res. 14, 6302–6309 (2008)
G.F. Guan, L.D. Wu, A.A.S. Bhagat, Z.R. Li, P.C.Y. Chen, S.Z. Chao, C.J. Ong, J.Y. Han, Sci. Rep. 3, 1475 (2013)
G.E. Hvichia, Z. Parveen, C. Wagner, M. Janning, J. Quidde, A. Stein, V. Muller, S. Loges, R.P.L. Neves, N.H. Stoecklein, H. Wikman, S. Riethdorf, K. Pantel, T.M. Gorges, Int. J. Cancer 138, 2894–2904 (2016)
K.A. Hyun, J. Kim, H. Gwak, H.I. Jung, Analyst 141, 382–392 (2016)
C. Jin, S.M. McFaul, S.P. Duffy, X. Deng, P. Tavassoli, P.C. Black, H. Ma, Lab Chip 14, 32–44 (2014)
J.H. Kang, S. Krause, H. Tobin, A. Mammoto, M. Kanapathipillai, D.E. Ingber, Lab Chip 12, 2175–2181 (2012)
N.M. Karabacak, P.S. Spuhler, F. Fachin, E.J. Lim, V. Pai, E. Ozkumur, J.M. Martel, N. Kojic, K. Smith, P.I. Chen, J. Yang, H. Hwang, B. Morgan, J. Trautwein, T.A. Barber, S.L. Stott, S. Maheswaran, R. Kapur, D.A. Haber, M. Toner, Nat. Protoc. 9, 694–710 (2014)
B.L. Khoo, M.E. Warkiani, D.S.W. Tan, A.A.S. Bhagat, D. Irwin, D.P. Lau, A.S.T. Lim, K.H. Lim, S.S. Krisna, W.T. Lim, Y.S. Yap, S.C. Lee, R.A. Soo, J. Han, C.T. Lim, PLoS One 9, e99409 (2014)
M.S. Kim, T.S. Sim, Y.J. Kim, S.S. Kim, H. Jeong, J.M. Park, H.S. Moon, S.I. Kim, O. Gurel, S.S. Lee, J.G. Lee, J.C. Park, Lab Chip 12, 2874–2880 (2012)
Y.J. Kim, S.H. Kim, T. Fujii, Y.T. Matsunaga, Biomater. Sci. 4, 953–957 (2016)
P. Li, Z.M. Mao, Z.L. Peng, L.L. Zhou, Y.C. Chen, P.H. Huang, C.I. Truica, J.J. Drabick, W.S. El-Deiry, M. Dao, S. Suresh, T.J. Huang, Proc. Natl. Acad. Sci. U. S. A. 112, 4970–4975 (2015)
C.J. Liao, C.H. Hsieh, H.M. Wang, W.-P. Chou, T.K. Chiu, J.H. Chang, A.C. Chao, M.H. Wu, RSC Adv. 7, 29339–29349 (2017)
L.S. Lim, M. Hu, M.C. Huang, W.C. Cheong, A.T.L. Gan, X.L. Looi, S.M. Leong, E.S.C. Koay, M.H. Li, Lab Chip 12, 4388–4396 (2012)
W. Liu, L. Nie, F. Li, Z.P. Aguilar, H. Xu, Y. Xiong, F. Fu, H. Xu, Biomater. Sci. 4, 159–166 (2016)
S. Nagrath, L.V. Sequist, S. Maheswaran, D.W. Bell, D. Irimia, L. Ulkus, M.R. Smith, E.L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U.J. Balis, R.G. Tompkins, D.A. Haber, M. Toner, Nature 450, 1235–U1210 (2007)
E. Ozkumur, A.M. Shah, J.C. Ciciliano, B.L. Emmink, D.T. Miyamoto, E. Brachtel, M. Yu, P.I. Chen, B. Morgan, J. Trautwein, A. Kimura, S. Sengupta, S.L. Stott, N.M. Karabacak, T.A. Barber, J.R. Walsh, K. Smith, P.S. Spuhler, J.P. Sullivan, R.J. Lee, D.T. Ting, X. Luo, A.T. Shaw, A. Bardia, L.V. Sequist, D.N. Louis, S. Maheswaran, R. Kapur, D.A. Haber, M. Toner, Sci. Transl. Med. 5, 179ra47 (2013)
P. Patil, T.K. Madhuprasad, D. Losic, M. Kurkuri, RSC Adv. 5, 89745–89762 (2015)
S. Riethdorf, H. Fritsche, V. Mueller, T. Rau, C. Schindibeck, B. Rack, W. Janni, C. Coith, K. Beck, F. Jaenicke, S. Jackson, T. Gornet, M. Cristofanilli, K. Pantel, Clin. Cancer Res. 13, 920–928 (2007)
Y. Song, T. Tian, Y. Shi, W. Liu, Y. Zou, T. Khajvand, S. Wang, Z. Zhu, C. Yang, Chem. Sci. 8, 1736–1751 (2017)
S.L. Stott, C.H. Hsu, D.I. Tsukrov, M. Yu, D.T. Miyamoto, B.A. Waltman, S.M. Rothenberg, A.M. Shah, M.E. Smas, G.K. Korir, F.P. Floyd, A.J. Gilman, J.B. Lord, D. Winokur, S. Springer, D. Irimia, S. Nagrath, L.V. Sequist, R.J. Lee, K.J. Isselbacher, S. Maheswaran, D.A. Haber, M. Toner, Proc. Natl. Acad. Sci. U. S. A. 107, 18392–18397 (2010)
Z. Wang, W. Wu, Z. Wang, Y. Tang, Y. Deng, L. Xu, J. Tian, Q. Shi, Analyst 141, 3621–3625 (2016)
M.E. Warkiani, B.L. Khoo, D.S.W. Tan, A.A.S. Bhagat, W.-T. Lim, Y.S. Yap, S.C. Lee, R.A. Soo, J. Han, C.T. Lim, Analyst 139, 3245–3255 (2014)
G. Xu, Y. Tan, T. Xu, D. Yin, M. Wang, M. Shen, X. Chen, X. Shi, X. Zhu, Biomater. Sci. 5, 752–761 (2017)
L. Yu, S.R. Ng, Y. Xu, H. Dong, Y.J. Wang, C.M. Li, Lab Chip 13, 3163–3182 (2013)
W. Zhang, S. Guo, W.S. Pereira Carvalho, Y. Jiang, M.J. Serpe, Anal. Methods 8, 7847–7867 (2016)
M.X. Zhao, P.G. Schiro, J.S. Kuo, K.M. Koehler, D.E. Sabath, V. Popov, Q.H. Feng, D.T. Chiu, Anal. Chem. 85, 2465–2471 (2013)
Acknowledgements
This research was supported by the National High-tech R&D Program of China (863 Program 2015AA020401), the National Basic Research Program (2015CB931801) and the National Natural Science Foundation of China (51690151, 51473093).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Jin, X., Chen, R., Zhao, S. et al. An efficient method for CTCs screening with excellent operability by integrating Parsortix™-like cell separation chip and selective size amplification. Biomed Microdevices 20, 51 (2018). https://doi.org/10.1007/s10544-018-0293-5
Published:
DOI: https://doi.org/10.1007/s10544-018-0293-5