Abstract
Rotifers are small, ubiquitous invertebrate animals found throughout the world and have emerged as a promising model system for studying molecular mechanisms in the fields of experimental ecology, aquatic toxicology, and geroscience. However, the lack of efficient gene expression manipulation techniques has hindered the study of rotifers. In this study, we used the L4440 plasmid with two reverse-oriented T7 promoters, along with RNase-deficient E. coli HT115, to efficiently produce dsRNA and thereby present an efficient feeding-based RNAi method in Brachionus plicatilis. We targeted Bp-Ku70 & Ku80, key proteins in the DNA double-strand breaks repair pathway, and then subjected rotifers to UV radiation. We found that the mRNA expression, fecundity, as well as survival rate diminished significantly as a result of RNAi. Overall, our results demonstrate that the feeding-based RNAi method is a simple and efficient tool for gene knockdown in B. plicatilis, advancing their use as a model organism for biological research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene expression manipulation techniques to achieve over-expression or knockdown are essential to explore gene functions in organisms. RNA interference (RNAi) has boosted several fields of biology by enabling study of loss-of-function effects in various organisms without laborious procedures or sophisticated apparatus (Ahringer 2006; Fire et al. 1998). RNAi is an evolutionally conserved post-transcriptional gene silencing mechanism, which double-strand RNA (dsRNA) molecules trigger degradation of the mRNA complementary to one strand of the dsRNA (Elbashir et al. 2001a, 2001b). This mechanism of gene silencing has led to universal use of reverse genetics methods and a better understanding of gene function at molecular levels.
Brachionus rotifer is a ubiquitous zooplankton and valuable for toxicology, evolutionary biology, gerontology and ecology (Gribble and Mark Welch 2017; Snell and Hicks 2011; Snell and Johnston 2014; Stelzer et al. 2021; Zhang et al. 2023). It has emerged as a model for experimental ecology to understand how changing environmental conditions influence gene functions and, in turn, how individuals and populations cope with changing environments (Franch-Gras et al. 2018; Kan et al. 2023; Kim et al. 2011; Lee et al. 2020). In order to fully realize Brachionus rotifer for molecular studies, techniques for experimental genetic manipulation are essential. Methodology for gene knockout based on CRISPR-Cas9 via microinjection is now available in rotifer, but it requires advanced microscopes, micromanipulators, special model of micropipette puller, and experienced technicians (Feng et al. 2023). Additionally, knockout of some genes can exhibit lethality. Hence, RNAi maybe a more universal and flexible method for rotifer’s gene function research. To our knowledge, there is only one technique described for RNAi in Brachionus rotifer, which involves transfecting small dsRNA into the hatched rotifers with lipofection agent (Shearer and Snell 2007). Although this method has allowed for the study of gene functions in rotifer, there are some drawbacks and limitations. Direct chemical synthesis or in vitro transcription of dsRNA is an expensive approach, producing limited quantities each time. As a result, experiments can only be conducted on a small number of organisms, which severely limits the experimental scale. Moreover, RNA is easily degraded, which limits the duration and efficiency of gene knockdown. Thus, a more efficient and economical protocol is needed for rotifer research.
In nematode Caenorhabditis elegans, RNAi was carried out by injection or soaking with dsRNA solution, or feeding the worms with bacteria expressing specific dsRNA (Ahringer 2006). The former is similar with transfection and suitable for treating a moderately number of organisms (e.g., tens to hundreds) or for high throughput screening in 96 well format. The latter method, involving feeding, is the least labor-intensive and most cost-effective, making it suitable for treating large quantities of organisms simultaneously. This approach has been successfully adopted in various animals, including crickets, planarians, and termites (La Fauce and Owens 2013; Newmark et al. 2003; Zhou et al. 2008). Given the presence of proteins essential for RNAi in the rotifer genome, such as the transmembrane protein SID-1, the RNA nucleases Dicer, and the Argonaute proteins, we infer that feeding RNAi should be functional in rotifers. If the RNAi feeding method is successful in rotifers, it would profoundly advance the development of rotifer research. In this study, we show the method for efficient gene knockdown in Brachionus rotifer by feeding RNAi.
UV-B radiation is known to have negative effects on aquatic animals, raising the mortality rate and harming fecundity (Hader et al. 2007; Kouwenberg et al. 1999; Zhu et al. 2021). At molecular scale, UV-B was found to cause DNA double-strand breaks (DSBs) and increase expression of DNA repair genes (e.g. Rad-51, LIG-3, Ku70 & Ku80) (Kan et al. 2023). We employed Brachionus plicatilis in our study and targeted the Ku70 and Ku80 genes, which encode proteins essential for repairing DSBs, for RNAi (Chapman et al. 2012; Ciccia and Elledge 2010; Lieber 2010). This choice was based on the easily identifiable changes in life table parameters that would occur if the RNAi was successful under UV radiation. Our results provide evidences for establishing of an easy, feeding-based RNAi method in Brachionus rotifers. We demonstrate reduced Ku70 and Ku80 mRNA, diminished the total fecundity, and shortened lifespan in response to UV radiation in Brachionus rotifer that fed on bacteria that express a dsRNA specific for Ku70 and Ku80. In summary, this method further promotes rotifers as a model for biological research.
Materials and methods
Rotifer and algae culture
The B. plicatilis strain JPNAG001 was cultivated in artificial seawater (ASW) with a salinity of 30 parts per thousand (ppt), prepared using concentrated sea salt (Blue Grand Star, China). The clone strain was maintained in an illuminated incubator set to a light intensity of 4000 lx, with a 12-h light:12-h dark photoperiod, at a temperature of 25 °C. The culture method for the rotifers was based on Snell’s methods with minor adjustments (Snell et al. 2012). The animal rights are not applicable.
The algae Chlorella vulgaris were cultured in F/2 medium at 25 °C, under continuous light exposure of 4000 lx (Guillard 1975). The algae were harvested at the exponential growth phase using centrifugation at 5000 rpm for 10 min. The rotifers were fed once daily with a concentrated suspension of C. vulgaris at a density of 3 × 106 cells/mL.
Gene idenfication and analysis
In brief, hidden Markov models for searching proteins with conservative domains (dsRNA-gated channel SID-1 family: PF13965; Dicer dimerisation domain: PF03368; Dicer dsRNA-binding domain: PF20932; Piwi domain: PF02171; Argonaute PAZ domain: PF18309) were obtained from the Pfam database (https://www.ebi.ac.uk/interpro/entry/pfam/#table) (Bateman et al. 2000). The genomes of B. plicatilis (NCBI GenBank: GCA_003710015.1) were scanned by the online tool ‘hmmsearch’ and keep significance E-values as 0.01 (https://www.ebi.ac.uk/Tools/hmmer/search/hmmsearch) (Finn et al. 2011; Franch-Gras et al. 2018; Potter et al. 2018). Sequence alignment was completed by ‘MUSCLE’ tool using MEGA 11 software (Edgar 2004; Tamura et al. 2021). The phylogenetic trees based on the Maximum-Likelihood method were performed by IQ-TREE (https://www.hiv.lanl.gov/content/sequence/IQTREE/iqtree.html) (Trifinopoulos et al. 2016). Conserved domains were scanned by ‘Batch CD-search’ tool and the E-value threshold was kept as default (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (Marchler-Bauer et al. 2017).
Synchronization of rotifers
Well-fed rotifers were collected with 300-mesh sieve and transferred into a 1.5 mL centrifuge tube. A 1000 μL pipette with micropipette tip was then used to pipetting rotifers forcefully. Finally, amictic eggs were collected and transferred to ASW containing C. vulgaris as a food source and left overnight to hatch (Feng et al. 2023). The next day, age-synchronized neonates were employed in subsequent experiments.
Vector and expression system
The RNAi feeding assay contains two key technique points, a kind of vector containing two T7 promotes in opposite orientations (L4440) and a RNase III deficient DE3 bacteria strain (HT115). DE3 E. coli have been integrated T7 RNA polymerase and can be induced with IPTG (isopropyl-β-D-thiogalactoside) to produce T7 RNA polymerase. This promises the production of dsRNA from DNA fragment cloned between the two T7 promoters. L4440 plasimd was obtained from Addgene (#1654, gift from Dr. Andrew Fire to Addgene). HT115 DE3 E. coli strain was obtained from Huayueyang (NRR10220, Beijing) (Dasgupta et al. 1998).
Target genes and plasmid constructs
In this study, we target Ku70 and Ku80 transcripts for RNAi (Table S1). Total RNA extraction and the first-strand cDNA synthesis were both completed by commercial kit (TIANGEN DP451 & Beyotime D7170M with gDNA eraser). The cDNA was used as PCR template and each construct was subcloned into pMD-19T (Takara, Japan) and sequenced for identification with commercial primers (e.g. M13 Primers). Appropriate restriction enzyme cutting sites were added to the 5’ end of primers for cloning. Subsequently, the target fragments and L4440 vectors were cut off by double restriction endonucleases simultaneously and then connected by T4 ligase (Takara, Japan). The recombinant plasmids were mixed with competent E. coli HT115 and transfected by thermal shock method. More detailed procedures can be found in the supplementary material (Feeding RNAi protocol). The primers used in this study were listed in Table 1.
Feeding protocol
Bacteria carrying the recombinant plasmid were added into 400 mL fresh Luria Broth (LB) medium and culture at 37 °C with shaking at 200 rpm until the optical density (OD600) reached approximately 0.6. IPTG was then added to a final concentration of 0.5 mM to induce plasmid transcription. After 8 h of induction, the bacteria were harvested by centrifugation at 5000 rpm for 10 min, resuspended in 100 mL of double-distilled H2O (ddH2O), and stored at 4 °C. Total RNA was extracted from the induced bacteria and subsequently mixed with RNase A in a high-salt concentration environment (NaCl ≥ 300 mM) to digest the single-stranded RNA. This treatment allowed for the assessment of dsRNA induction efficiency via 1% agarose gel electrophoresis.
Approximately 5000 one-day-old rotifers were carefully transferred into a 50 mL beaker containing culture medium. The medium was composed of 3 × 106 cells/mL of C. vulgaris suspended in 30 ppt ASW and supplemented with 20 μM 5’-Deoxy-5-fluorouridine (FUdR), which inhibits the hatching of new born eggs (Snell et al. 2012). For the RNAi treatment, the volume of induced bacteria added was one-tenth of the total medium volume, resulting in a concentration of approximately 1 × 107 cells/mL. The RNAi treatment duration was 48 h, during which the culture medium was refreshed daily. The efficiency of RNAi was assessed using RT-qPCR, with the empty vector serving as positive control.
UV treatment and life table experiment
In the experiment targeting the knockdown of Ku70 and Ku80, UV-B radiation was employed to induce DNA DSBs. The UV-B radiation intensity was set at 1 W/m2, as measured by a UV-B radiometer (Beijing Normal University Optoelectronic Technology). The radiation dose was administered at a rate of 1 kJ/m2 per day by hanging three UV-B lamps (Sankyo Denki, G8T5E, Japan) approximately 20 cm above the culture plates for a duration of 60 min. The UV-B lamps were covered with cellulose diacetate filters (Chengdu First Instrument) to eliminate shortwave radiation below 290 nm, thereby simulating a spectral more akin to natural sunlight (Teramura et al. 2006). To ensure UV-B penetration and to prevent evaporation during the exposure period, the culture plates were sealed with a quartz lid (Kim et al. 2011). A total of 48 newborn rotifers were individually transferred into 24-well culture plates, each well containing two rotifers and 1 mL of the previously described culture medium, which was supplemented with the induced bacteria. The medium was refreshed every day, and daily inspections were conducted to monitor the survival of the rotifers and the production of offspring until all individuals were dead.
Quantitative real-time PCR (qPCR)
Approximately 5000 rotifers were processed into qPCR. Total RNA was extracted using TIANGEN commercial kit (DP-451, Beijing, China) and 1 μg of total RNA was converted to cDNA by primers mixture containing poly(T) and random primers (Beyotime D7170M, Shanghai, China). qPCR was performed by using SYBR Green Premix (SinoMol SQ202–01, Nanjing, China). Data was collected as threshold cycle (CT) values and β-actin was used as the housekeeping gene. Expression levels of target genes were obtained by normalizing them to housekeeping genes, and the ΔΔCt method was used to calculate fold changes in relative gene expression (Livak and Schmittgen 2001). Primers used were listed in Table 1. The qPCR thermocycling was as follows: Predenaturation at 95 °C for 60 s, 40 cycles of denaturation at 95 °C for 15 s, and annealing and extension at 60 °C for 60 s. Melting curve steps: 95 °C for 15 s, 60 °C for 60 s, 95 °C for 15 s and continuous (Roche, Light Cycler 96).
Statistical analysis
Statistical analysis was conducted by GraphPad Prism Software. Student’s t-test was applied when two data sets were compared. The comparisons between groups were analyzed by one-way ANOVA, and a multiple-comparison Tukey’s test was used to determine significant differences between each group and the control. Data were presented as means ± SD. P-values < 0.05 were considered statistically significant.
Results
Core players of RNAi in Brachionus rotifer
The rotifer B. plicatilis carries all the key components necessary for RNAi within its genome (Fig. 1). The SID-1 gene (systemic RNAi defective), which is crucial for systemic RNAi, encodes a transmembrane protein that forms a channel for dsRNA gating. Through genomic analysis of B. plicatilis, we identified a homolog of the SID-1 gene. Additionally, we found that the genome of B.plicatilis contains two homologs of Dicer and eleven homologs of Argonaute proteins, which are vital for RNA processing and mRNA degradation, respectively. Given the presence of these essential genes for RNAi, we infer that RNAi mechanism induced by feeding is likely to be functional in rotifers.
Players of RNA interference in Brachionus plicatilis. a The domain organization of SID-1 and Dicer protein in rotifer, nematode and fruit fly. Brachionus plicatilis (B.p) SID-1 (RNA32255.1), Dicer-1 (RNA31321.1), Dicer-2 (RNA38511.1); Caenorhabditis elegans (C.e) SID-1 (AAL78657.1), Dicer (NP_498761.2); Drosophila melanogaster (D.m) Dicer-1 (NP_524453.1), Dicer-2 (NP_523778.2). Accession numbers are for Genbank. b The phylogenetic tree of Argonaute proteins based on the Maximum-Likelihood method. Argonaute can be divided into Ago family and Piwi family. The accession numbers for Genbank are listed after the taxon name. Shizosaccharomyces pombe (S.p), Arabidopsis thaliana (A.t), Brachionus plicatilis (B.p), Drosophila melanogaster (D.m), Caenorhabditis elegans (C.e), Homo sapiens (H.s). c The Agronaure protein is highly conserved between Brachionus plicatilis and Homo sapiens. Brachionus plicatilis (B.p) Ago-1 (RNA31902.1); Homo sapiens (H.s) Ago-1 (NP_036331.1)
PCR and dsRNA induction
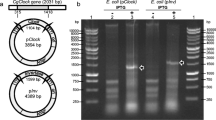
Specific primers were designed to amplify an approximately 550 base pair segments from the Bp-Ku70 or Bp-Ku80 genes, which was subsequently sub-cloned into the L4440 plasmid (Fig. 2a and Table S1). To confirm the production of the target double-stranded RNA, the total RNA extracted from IPTG-induced bacteria was digested and analyzed. Figure 2c illustrates that distinct bands corresponding to the expected size of the dsRNA are evident in the IPTG-induced samples, whereas no dsRNA expression is observed in the uninduced control bacteria.
Polymerase chain reaction and induction of double-strand RNA of Ku70 and Ku80. a Schematic diagram of target gene and amplification position. The size of amplifications is about 550 bp. b Schematic diagram of the plasmid L4440. MCS means multiple cloning sites. c The gel electrophoresis image on the left shows PCR products, while the image on the right illustrates bacterial total RNA digested by RNase A. The target bands are marked with red boxes or an arrow
Optimization of RNAi feeding protocol
To standardize the RNAi feeding assay, we fixed the feeding amount of induced bacteria (approximately 1 × 107 cells/mL) and evaluated the interference efficiency under a range of feeding durations (Fig. 3a). The feeding regimen was systematically varied from one to four days, and mRNA levels were quantified using qPCR. As the feeding duration was increased, the mRNA levels exhibited a significant decline and stabilized after two days of feeding (Fig. 3b). Consequently, for the RNA interference method relying on feeding in rotifer, we recommend maintaining the interference duration at two days or longer, with the specific duration determined by the experimental needs.
The relative expression of Ku70 changes with the duration of RNA interference. a Schematic diagram of feeding RNAi workflow. b By the second day of continuous interference, a significant decrease in relative expression was observed, and continued interference maintained the reduced mRNA level. Bacteria carrying an empty plasmid were taken as a control. Data are presented as mean ± SD, n = 3. F4, 10 = 46.09, P < 0.0001. Tukey post-hoc, a > b, α = 0.05
Knocking down Ku70 and Ku80 makes rotifers sensitive to UV radiation
To evaluate the effectiveness of RNA interference induced by feeding, we supplied B. plicatilis with bacteria expressing dsRNA targeting Ku70 and Ku80, which are pivotal genes in the DNA double-strand breaks repair pathway following UV irradiation. Compared to the control group, after UV radiation, the fecundity of rotifers with Ku70 and Ku80 gene knockdown significantly decreased by 58.53%, and their lifespan was also reduced by 26.40% (Fig. 4b, c). Moreover, we also detected that the knockdown of Ku70 and Ku80 could result in the impairment of embryonic development in certain cases (Fig. 4d). These findings collectively confirm that our RNAi method is effective in inducing targeted gene silencing in rotifers.
Knocking down Ku70 & Ku80 makes rotifer fragile to UV radiation. a qPCR was used to detect knockdown efficiency. The mRNA level of Ku70 & Ku80 significantly reduced after RNAi feeding regimen. Bacteria carrying an empty plasmid were taken as a control. Data are presented as mean ± SD, n = 3. (Ku70) t = 11.19, df = 14, P < 0.0001 (two-tailed), a > b, α = 0.05; (Ku80) t = 9.414, df = 14, P < 0.0001 (two-tailed), a > b, α = 0.05. b Survival curve analysis for Ku70 & Ku80 knockdown rotifers after UV radiation. Bacteria carrying an empty plasmid were taken as a control. Data are analysed by Log-rank test (Mantel-Cox). c The fecundity reduced significantly after RNAi feeding regimen. Bacteria carrying an empty plasmid were taken as a control. Data are presented as mean ± SD, n = 3. t = 12.87, df = 70, P < 0.0001 (two-tailed), a > b, α = 0.05. d Abnormal embryonic development was observed in rotifers subjected to co-RNAi targeting Ku70 and Ku80. Scale bar: 50 μm
Discussion
RNA interference (RNAi) is a powerful technique that allows for the selective suppression of gene expression, thereby enabling the investigation of gene function (Fire et al. 1998). Traditionally, in animal systems, RNAi has been achieved through methods such as injection or transfection (Ahringer 2006). While these approaches have demonstrated varying degrees of success, they are not without limitations. Injecting double-stranded RNA into small zooplankton, such as rotifers, presents significant challenges due to the difficulty in operation and the potential for mechanical damage, which can lead to developmental abnormalities and accidental mortality in the animals (Feng et al. 2023). Moreover, the production of dsRNA is often limited, and the scale of experiments can be small, posing constraints on the transfection method (Ahringer 2006; Shearer and Snell 2007). In the present study, we employed a feeding-based approach to deliver dsRNA, which offers advantages such as labor efficiency, cost-effectiveness, and the ability to conduct experiments on a large scale with a multitude of individuals. This method has been successful in inducing targeted gene silencing in this study.
The RNA interference is a conserved mechanism across eukaryotes and is initiated by the presence of double-stranded RNA (Elbashir et al. 2001a, 2001b). The dsRNA is initially transported into the cell through the Sid-1 transmembrane protein, where it is processed by the Dicer enzyme into 21 ~ 23 nucleotide small RNA duplexes known as siRNAs (Feinberg and Hunter 2003; Shih and Hunter 2011; Winston et al. 2002; Zhang et al. 2004). These siRNAs then associate with Argonaute proteins to form RNA-induced silencing complexes (RISCs), which function to target and degrade mRNA that is complementary to the siRNA sequence, guided by base pairing principles (Jin et al. 2021; Liu et al. 2004; Sheu-Gruttadauria and MacRae 2017; Yuan et al. 2005). In the genome of B. plicatilis, we identified Sid-1, Dicer, and Argonaute proteins, each with homologs, indicating the presence of essential RNAi components (Fig. 1). Sid-1 is widely conserved and facilitates the transport of RNAi silencing signals between cells (Feinberg and Hunter 2003; Shih and Hunter 2011; Winston et al. 2002). It is found in various invertebrates, including Brachionus rotifers. Dicer proteins, initially characterized in humans, possess a complex domain structure, which typically includes a DEAD box domain, an RNA helicase domain, a dimerization domain, a PAZ domain, two RNase III domains, and a dsRNA binding domain (Höck and Meister 2008; Sasaki and Shimizu 2007). However, some Dicer homologs may lack certain domains. For instance, the DEAD domain, crucial for the regulation of various RNA-related processes, is notably absent in Bp-Dicer2. This domain loss is also commonly observed in mollusks, annelids, platyhelminths, and most arthropods (Rossi et al. 2014). Notwithstanding the absence of the DEAD domain in some Dicer homologs, the other domains of Dicer proteins in Brachionus rotifers remain conserved. This includes the PAZ (Piwi-Argonaute-Zwille) domain, which is essential for identifying the ends of double-stranded RNA (dsRNA), and the RNase III domain, responsible for cleaving dsRNA (Park et al. 2011; Sasaki and Shimizu 2007; Zhang et al. 2004). Argonaute proteins, on the other hand, can be categorized into the Ago and Piwi families. The Ago family is expressed ubiquitously, whereas the Piwi family is specifically expressed in the germline. Both families share the PAZ domain with Dicer and are distinguished by the presence of a unique Piwi domain, which is responsible for binding siRNAs and cleaving mRNA (Höck and Meister 2008; Sheu-Gruttadauria and MacRae 2017; Yuan et al. 2005). The key amino acid residues for dsRNA binding and the cleavage active sites are shown in the figure S1. This conservation of sequence and structural information further corroborates that feeding RNAi may function well in rotifers.
The Ku protein complex is capable of recognizing the ends of DNA double-strand breaks and, upon binding to DNA, recruits and activates the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), thereby initiating the nonhomologous end joining (NHEJ) pathway for DNA damage repair (Chapman et al. 2012; Ciccia and Elledge 2010; Lieber 2010). Our research indicates that RNAi in Brachionus rotifers not only results in a significant reduction in mRNA levels but also leads to a pronounced decrease in fecundity and an increase in mortality under UV radiation (Fig. 4b, c). It is important to note that RNAi is a post-transcriptional regulatory mechanism that degrades mRNA without directly affecting mRNA transcription, thus its effects are transient. Our experiments revealed that a sustained feeding regimen of two days or longer was required to maintain stable mRNA levels. Additionally, we found that the reduction of Ku70 and Ku80 transcripts in some rotifers led to abnormal embryonic development (Fig. 4d), suggesting that RNAi in rotifers may be systemic and can impact the germline, offering a potential avenue for investigating interference during rotifer embryonic development.
In any case, these findings underscore the effectiveness of RNA interference and the feeding method in Brachionus rotifer. This approach represents a valuable tool for genetic manipulation of this organism in the fields of aquatic toxicology, evolutionary biology, biogerontology, and experimental ecology.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Ahringer J (2006) Reverse genetics. WormBook. https://doi.org/10.1895/wormbook.1.47.1
Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL (2000) The Pfam protein families database. Nucleic Acids Res 28(1):263–266. https://doi.org/10.1093/nar/28.1.263
Chapman JR, Taylor MR, Boulton SJ (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47(4):497–510. https://doi.org/10.1016/j.molcel.2012.07.029
Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40(2):179–204. https://doi.org/10.1016/j.molcel.2010.09.019
Dasgupta S, Fernandez L, Kameyama L, Inada T, Nakamura Y, Pappas A, Court DL (1998) Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III — the effect of dsRNA binding on gene expression. Mol Microbiol 28(3):629–640. https://doi.org/10.1046/j.1365-2958.1998.00828.x
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001a) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836):494–498. https://doi.org/10.1038/35078107
Elbashir SM, Lendeckel W, Tuschl T (2001b) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15(2):188–200. https://doi.org/10.1101/gad.862301
Feinberg EH, Hunter CP (2003) Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301(5639):1545–1547. https://doi.org/10.1126/science.1087117
Feng HY, Bavister G, Gribble KE, Welch DM (2023) Highly efficient CRISPR-mediated gene editing in a rotifer. PLoS Biol. https://doi.org/10.1371/journal.pbio.3001888
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39(suppl):W29–W37. https://doi.org/10.1093/nar/gkr367
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811. https://doi.org/10.1038/35888
Franch-Gras L, Hahn C, Garcia-Roger EM, Carmona MJ, Serra M, Gomez A (2018) Genomic signatures of local adaptation to the degree of environmental predictability in rotifers. Sci Rep 8(1):16051. https://doi.org/10.1038/s41598-018-34188-y
Gribble KE, Mark Welch DB (2017) Genome-wide transcriptomics of aging in the rotifer Brachionus manjavacas, an emerging model system. BMC Genomics 18(1):217. https://doi.org/10.1186/s12864-017-3540-x
Guillard RRL (1975) Culture of Phytoplankton for Feeding Marine Invertebrates. pp 29–60. In Smith WL, Chanley MH (eds) Culture of Marine Invertebrate Animals. Springer, Boston, MA. https://doi.org/10.1007/978-1-4615-8714-9_3
Hader DP, Kumar HD, Smith RC, Worrest RC (2007) Effects of solar UV radiation on aquatic ecosystems and interactions with climate change. Photochem Photobiol Sci 6(3):267–285. https://doi.org/10.1039/b700020k
Höck J, Meister G (2008) The Argonaute protein family. Genome Biol. https://doi.org/10.1186/gb-2008-9-2-210
Jin S, Zhan J, Zhou Y (2021) Argonaute proteins: structures and their endonuclease activity. Mol Biol Rep 48(5):4837–4849. https://doi.org/10.1007/s11033-021-06476-w
Kan D, Zhang Y, Zeng J, Lian H, Feng L, Feng Y, Liu X, Han C, Yang J (2023) Physiological response and molecular mechanisms against UV-B radiation in Brachionus asplanchnoidis (Rotifera). Ecotoxicol Environ Saf. https://doi.org/10.1016/j.ecoenv.2023.115319
Kim RO, Rhee JS, Won EJ, Lee KW, Kang CM, Lee YM, Lee JS (2011) Ultraviolet B retards growth, induces oxidative stress, and modulates DNA repair-related gene and heat shock protein gene expression in the monogonont rotifer. Brachionus Sp Aquat Toxicol 101(3–4):529–539. https://doi.org/10.1016/j.aquatox.2010.12.005
Kouwenberg JHM, Browman HI, Cullen JJ, Davis RF, St-Pierre JF, Runge JA (1999) Biological weighting of ultraviolet (280-400 nm) induced mortality in marine zooplankton and fish. I. Atlantic cod (Gadus morhua) eggs. Marine Biol 134(2):269–284. https://doi.org/10.1007/s002270050545
La Fauce K, Owens L (2013) Suppression of Penaeus merguiensis densovirus following oral delivery of live bacteria expressing dsRNA in the house cricket (Acheta domesticus) model. J Invertebr Pathol 112(2):162–165. https://doi.org/10.1016/j.jip.2012.11.006
Lee YH, Kang HM, Kim MS, Lee JS, Wang M, Hagiwara A, Jeong CB, Lee JS (2020) Multigenerational mitigating effects of ocean acidification on in vivo endpoints, antioxidant defense, DNA damage response, and epigenetic modification in an asexual monogonont rotifer. Environ Sci Technol 54(13):7858–7869. https://doi.org/10.1021/acs.est.0c01438
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. https://doi.org/10.1146/annurev.biochem.052308.093131
Liu JD, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305(5689):1437–1441. https://doi.org/10.1126/science.1102513
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203. https://doi.org/10.1093/nar/gkw1129
Newmark PA, Reddien PW, Cebrià F, Alvarado AS (2003) Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. P Natl Acad Sci USA 100:11861–11865. https://doi.org/10.1073/pnas.1834205100
Park J-E, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN (2011) Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475(7355):201–205. https://doi.org/10.1038/nature10198
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD (2018) HMMER web server: 2018 update. Nucleic Acids Res 46(W1):W200–W204. https://doi.org/10.1093/nar/gky448
Rossi JJ, Gao Z, Wang M, Blair D, Zheng Y, Dou Y (2014) Phylogenetic analysis of the endoribonuclease dicer family. PLoS ONE. https://doi.org/10.1371/journal.pone.0095350
Sasaki T, Shimizu N (2007) Evolutionary conservation of a unique amino acid sequence in human DICER protein essential for binding to Argonaute family proteins. Gene 396(2):312–320. https://doi.org/10.1016/j.gene.2007.04.001
Shearer TL, Snell TW (2007) Transfection of siRNA into Brachionus plicatilis (Rotifera). Hydrobiologia 593(1):141–150. https://doi.org/10.1007/s10750-007-9067-4
Sheu-Gruttadauria J, MacRae IJ (2017) Structural foundations of RNA silencing by argonaute. J Mol Biol 429(17):2619–2639. https://doi.org/10.1016/j.jmb.2017.07.018
Shih JD, Hunter CP (2011) SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17(6):1057–1065. https://doi.org/10.1261/rna.2596511
Snell TW, Hicks DG (2011) Assessing toxicity of nanoparticles using Brachionus manjavacas (Rotifera). Environ Toxicol 26(2):146–152. https://doi.org/10.1002/tox.20538
Snell TW, Johnston RK (2014) Glycerol extends lifespan of Brachionus manjavacas (Rotifera) and protects against stressors. Exp Gerontol 57:47–56. https://doi.org/10.1016/j.exger.2014.05.005
Snell TW, Fields AM, Johnston RK (2012) Antioxidants can extend lifespan of Brachionus manjavacas (Rotifera), but only in a few combinations. Biogerontology 13(3):261–275. https://doi.org/10.1007/s10522-012-9371-x
Stelzer CP, Blommaert J, Waldvogel AM, Pichler M, Hecox-Lea B, Mark Welch DB (2021) Comparative analysis reveals within-population genome size variation in a rotifer is driven by large genomic elements with highly abundant satellite DNA repeat elements. BMC Biol 19(1):206. https://doi.org/10.1186/s12915-021-01134-w
Tamura K, Stecher G, Kumar S, Battistuzzi FU (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Teramura AH, Ziska LH, Sztein AE (2006) Changes in growth and photosynthetic capacity of rice with increased UV-B radiation. Physiol Plant 83(3):373–380. https://doi.org/10.1111/j.1399-3054.1991.tb00108.x
Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44(W1):W232–W235. https://doi.org/10.1093/nar/gkw256
Winston WM, Molodowitch C, Hunter CP (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295(5564):2456–2459. https://doi.org/10.1126/science.1068836
Yuan Y-R, Pei Y, Ma J-B, Kuryavyi V, Zhadina M, Meister G, Chen H-Y, Dauter Z, Tuschl T, Patel DJ (2005) Crystal Structure of A. aeolicus Argonaute, a Site-Specific DNA-Guided Endoribonuclease, Provides Insights into RISC-Mediated mRNA Cleavage. Mol Cell 19(3):405–419. https://doi.org/10.1016/j.molcel.2005.07.011
Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human dicer and bacterial RNase III. Cell 118(1):57–68. https://doi.org/10.1016/j.cell.2004.06.017
Zhang Y, Zhou Y, Kan D, Yang Y, Shen J, Han C, Liu X, Yang J (2023) m6A-mediated nonhomologous end joining (NHEJ) pathway regulates senescence in Brachionus plicatilis (Rotifera). Arch Gerontol Geriatr. https://doi.org/10.1016/j.archger.2023.104994
Zhou X, Wheeler MM, Oi FM, Scharf ME (2008) RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem Mol Biol 38(8):805–815. https://doi.org/10.1016/j.ibmb.2008.05.005
Zhu L, Huang R, Zhou L, Xi Y, Xiang X (2021) Responses of the ecological characteristics and antioxidant enzyme activities in Rotaria rotatoria to UV-B radiation. Hydrobiologia 848(20):4749–4761. https://doi.org/10.1007/s10750-021-04671-1
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Numbers: 31772458) and the Graduate Student Scientific Research Innovation Projects in Jiangsu Province (Grant Numbers: KYCX23-1748).
Author information
Authors and Affiliations
Contributions
Yu Zhang: conceptualization, writing- Original draft preparation, visualization. Dongqi Kan & Yang Zhou: conceptualization, methodology. Hairong Lian, Lingling Ge & Jing Shen: data curation, formal analysis. Cui Han, Zhongqi Dai & Yan Shi: Rotifer and algae culture. Xiaojie Liu: writing—Review & Editing. Jiaxin Yang: supervision, funding acquisition.
Corresponding author
Ethics declarations
Competing interest
No conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Kan, D., Zhou, Y. et al. Efficient RNA interference method by feeding in Brachionus plicatilis (Rotifera). Biotechnol Lett (2024). https://doi.org/10.1007/s10529-024-03524-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10529-024-03524-w