Abstract
Objectives
For the stress from fermenters, downstream processing equipment, and wastewater treatment to be alleviated, lowering salt-dependence in the ectoine synthesis process is of great significance in the moderately halotolerant Halomonas hydrothermalis Y2.
Results
In H. hydrothermalis Y2, the σ70- and σ38-controlled promoters of ectA are predicted to be involved in the osmotic regulation of ectoine synthesis. By substituting the ectA promoter with a promoter P265 that identified in the outer membrane pore protein E of H. hydrothermalis Y2, the salt dependence of ectoine synthesis was significantly decreased. In the 500-ml flask containing various NaCl contents, the engineered strain (p/Y2/△ectD/△doeA) showed a remarkably enhanced ability in ectoine synthesis, especially under lower saline stress. After a 36-h fed-batch fermentation in the 1-l fermenter, p/Y2/△ectD/△doeA synthesized 11.5 g ectoine l−1 in the presence of 60 g NaCl−1 l, with a high 0.32 g ectoine l−1 h−1 productivity, a specific productivity of 512.2 mg ectoine per g cell dry weight (CDW)−1, and an excretion ratio of 67 % ectoine.
Conclusions
As no impaired growth was observed in strain p/Y2/△ectD/△doeA while ectoine synthesis was increased, this promoter engineering strategy provides a practical protocol for lowering the salt-dependence of ectoine synthesis in this moderately halotolerant strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
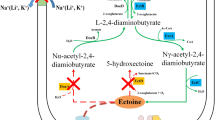
To cope with high osmotic stress in a saline environment, microorganisms developed two key mechanisms to achieve an osmotic balance between the cytoplasm and the surrounding medium (Oren 2002). Some halotolerant or halophiles following the organic osmolyte mechanism can amass compatible solutes to lower the potential of cytoplasmic water and to feed off the detrimental effects of high osmolality on cellular physiology and growth (Oren 2018). Recently, there has been compelling evidences for the bioactivities of these multifunctional molecules, which serve as osmostress protectants from a variety of abiotic stresses (Pejin et al. 2015; Jadhav et al. 2018). Among the known compatible solutes, the aspartate derivative ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid) is of great interest because of its broad applications in various fields, and several tons are produced annually via a biotechnological process by strain Halomonas elongata DSM2581T (Kunte 2006).
The production of ectoine is often strictly salt-dependent. However, the use of high-salinity media in such fermentation processes imposes notable constraints on the cost, design, and durability of fermenter systems, as well as saline wastewater treatment (Becker et al. 2013). To eliminate issues with high-salinity fermentation, considerable efforts have been made to explore alternative fermentation processes, including some engineering protocols for decreasing salinity requirements. For example, the heterologous pathway of ectoine synthesis was constructed in the E. coli and Corynebacterium glutamicum hosts to yield high ectoine production under low salinity (Becker et al. 2013; Seip et al. 2011). However, with respect to designing efficient chassis cells, there have been very limited studies on the modification of Halomonas species to decrease their dependence on high salt. In our recent publication, we constructed a mutant that lacks the Na+/H+ antiporter (Mrp) and showed that it retained similar ectoine productivity in a low-salinity medium (Zhao et al. 2019). This implies the engineering potentials of lowing the salt-dependence for ectoine production in this moderately halotolerant producer.
In H. elongata DSM2581T or most strains with putative ectoine synthesis pathways, the ectABC gene cluster, which is responsible for converting L-aspartate-β-semialdehyde to ectoine, is transcribed in different patterns (Calderon et al. 2004; Czech et al. 2018). In this study, the promoter assembly and the osmotic responses of ectABC in H. hydrothermalis Y2 were investigated. Based on the mutant Y2/△ectD/△doeA we previously constructed (Zhao et al. 2019), the ectA promoter region was replaced by a constitutive and strong promoter, and higher ectoine productivity was successfully obtained in a lower saline medium.
Materials and methods
Microorganisms, plasmids and culture conditions
The strains and plasmids used in this study are listed in Table 1. H. hydrothermalis Y2, a moderately halotolerant and alkalophilic strain, was isolated from alkaline pulp mill wastewater and was preserved in the China Center for Type Culture Collection (CCTCC No. M208188) (Yang et al. 2010). To screen for strong promoters in the H. hydrothermalis Y2 strain, promoters were ligated with the promoter probe vector pKK232-8 and transformed into Escherichia coli DH5α. The promoter replacement was based on strain Y2/ΔectD/ΔdoeA that we previously constructed (Zhao et al. 2019). E. coli strains and H. hydrothermalis Y2 strains were routinely maintained on Luria-Bertani (LB) agar plates and propagated in liquid LB medium containing (l−1) 10 g tryptone, 5 g yeast extract, and 10 g NaCl, pH 7.
Rapid amplification of cDNA ends polymerase chain reaction (RACE-PCR) and sequence alignment
Total RNA of strain Y2 was isolated from the exponentially growing cells using Easy Pure RNA Kit (TransGen Biotech, Beijing, China). To determine the transcriptional initiation sites, RACE-PCR was performed using the SMARTerTM RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA), and the primers ectA-F1 (GATTACGCCAAGCTTCGGGCCGATTCTTACCAGGTTTTCC) and ectB-F1 (GATTACGCCAAGCTTTGGCGCCCTGAGTGGGAATACCG). The transcription initiation site was defined by the boundary between the artificially attached RNA and the 5′-end of the transcript. In addition, the promoter sequences were also aligned by ClustalX2, with the corresponding regions from strains H. elongata DSM2581T and C. salexigens DSM3043T.
Transcriptome-based promoter screening
The transcriptome of H. hydrothermalis Y2 growing at various pH values (6.21, 8, and 10.17) were sequenced previously (Accession No: SRP075842) (Cheng et al. 2016). The potential strong promoters were screened by comparing the fragments per kilobase of transcript per million fragments mapped (FPKM) values of the corresponding genes under these conditions.
Genomic DNA of H. hydrothermalis Y2 was isolated using the ChargeSwitch® gDNA Mini Bacteria Kit (Invitrogen, Corp, Carlsbad, Calif., USA). The chloramphenicol acetyltransferase (CAT) ELISA kit100 (Roche) was used to evaluate the transcriptional activities of the screened promoters. Putative promoters were amplified using primers listed in Supplementary Table 1 and the genomic DNA of H. hydrothermalis Y2 as a template. The resulting fragments were then ligated with the promoter probe vector pkk232-8, transformed into E. coli DH5α competent cells, and spread on plates containing 25 mg chloramphenicol l−1. After cultivation at 30 °C, the positive clones were picked and sequenced for fidelity. For comparison, the ectA promoter was fused with vector pkk232-8, transformed into E. coli DH5α competent cells. In addition, the empty vector pkk232-8 was also transformed into E. coli DH5α and used as a negative control.
Promoter activity assay
For CAT activity measurement, the E. coli constructs were cultivated in the LB medium and collected by centrifugation at 8000×g for 5 min at 4 °C. After washed twice and re-suspended in 2 ml lysis buffer, cells were passed through a high-pressure homogenizer (JN-02C, JNBIO, China) and then centrifuged at 10,000×g for 10 min. The protein content of the supernatant was determined using Coomassie Brilliant Blue. The CAT activity was measured using an enzyme-linked immunoassay kit, and expression levels were characterized by the ratio of (differential dilution factor × CAT concentration)/(diluted fold × protein concentration).
To test the osmotic response of the ectA and ectB promoter regions, E. coli construct pkk-Y2-PectA fusing with the ectA promoter region was cultivated in LB media various NaCl contents, 0.5 M KCl, 1 M sucrose, and 1 M glycerol. After 10 h of cultivation, the CAT activities were assayed as described above.
Gene manipulation
For promoter replacement in the upstream of ectA, the recombinant fragment with P265 was constructed by overlap extension PCR (OE-PCR), using primers listed in Supplementary Table 2. The recombinant fragment was ligated with the shuttle vector pK18mobsacB and transferred into E. coli S17-1. The procedures for conjugation were described previously (Yang et al. 2010). Finally, the resulting strain was selected on LB medium containing 20 % (w/v) sucrose and verified by PCR and sequencing.
Ectoine production
To determine the osmotic stress response and ectoine production titers, strains Y2/△ectD/△doeA and p/Y2/△ectD/△doeA were cultivated in LBG medium (LB medium with 10 g glucose l−1 and 40 g NaCl l−1) for seed preparation, by cultivation at 30 °C for 8 h to their logarithmic phase. The cultures were then adjusted to a same OD value and diluted 200 fold into 500-ml baffled Erlenmeyer flasks containing 50 ml fresh MMG medium (Zhao et al. 2019), as well as 20, 40, 60, 80, or 100 g NaCl l−1. After incubating at 30 °C for 24 h, cultures were sampled for measuring the cell growth and ectoine production.
For fed-batch fermentation, the seed culture of p/Y2/△ectD/△doeA was prepared as described above and transferred into a 1-l Multifors fermenter (Infors HT) with initial NaCl, glucose, and mono sodium glutamate (MSG) concentrations (l−1) of 20 g, 5 g, and 20 g, respectively. The final NaCl concentration was 60 g l−1 by feeding NaCl solutions at 12, 15, and 18 h. The concentration of MSG was detected by SBA-40D (Shandong Academy of Sciences) every 3 h and was maintained at 20 (± 5) g l−1 by addition of 50 % MSG during the first 24-h fermentation.
Analysis methods
Growth was monitored by measuring the optical density at 600 nm and the cell dry weight (CDW). For intracellular ectoine analysis, the fermentation cultures were sampled and cells were collected by centrifugation at 6000×g for 10 min. After washed twice with 1.5 M NaCl solution, the cell pellets were resuspended in the distilled water and passed through the French Press (JN-02C, JNBIO, China) to obtain a cell homogenate. After further centrifugation at 8000×g for 10 min, two volumes ethanol were added to the supernant and mixed as the intracellular samples. For extracellular ectoine analysis, the culture was also centrifuged at 6000×g for 10 min, and the supernant was supplemented with two volumes ethanol and prepared as the extracellular samples. Before High Performance Liquid Chromatography (HPLC) analyses, the mixtures were filtered through a 0.25 µm microporous membrane. The Shimadzu HPLC system (Kyoto, Japan) that equipped with a Venusil XBP NH2 column (4.6 × 250 mm) was used, by applying the acetonitrile/H2O (60:40, v/v) as the mobile phase, at a flow rate of 0.5 ml min−1. The absorption of ectoine was recorded at 204 nm using an UV-detector.
Accession numbers
Sequences encoding various promoters in this study were extracted from the genomic sequence of strain Y2 (NCBI no: CP023656). The transcriptional data for promoter screening were extracted from the NCBI database of Accession No SRP075842.
Results
Promoter identification of ectABC genes by RACE-PCR
Multiple sequences alignment among the promoter sequences of ectA and ectB in strains Y2, H. elongata, and C. salexigens were performed by ClustalX2 (Fig. 1). In combination with the online prediction, the consensus sequences of AGCGAA in the upstream of ectA gene were proposed as a putative ribosome-binding site (RBS) that is conserved throughout prokaryotes. When compared with the identified promoters of H. elongata 2058T and C. salixigens 3043T, 5′-end RACE-PCR revealed a different promoter assembly for the ectoine synthesis of strain Y2. As shown in Fig. 2, two transcriptional initiation sites were mapped upstream of ectA and one was mapped in front of ectB. Based on the two mapped transcription initiation sites of ectA and the conservative sequences of typical promoters, one σ70- and one σ38-type promoters similar to those of H. elongata DSM2581T were identified; however, the σ38 promoter is located at a different location (Fig. 2a) (Schwibbert et al. 2011). Sequences of the − 10 (TATAAT) and − 35 (TTGAAT) regions of putative promoter PectA1 matched well with the consensus sequence of σ70 in E. coli (TATAAT-16–18 bp-TTGACA) (Wösten 1998), with only two base variations in the − 35 region. In addition, a 17-bp region linking the − 10 and − 35 regions were also found, which is typical for σ70 promoters. The promoter sequence of PectA2 is not conserved among two compared strains, but resembles the typical sequence of σ38-controlled promoters (Lee and Gralla 2004), with a well-conserved − 10 region (TAGACT) and a cytosine at the − 13 position. In accordance with previous observations that the − 35 region of σ38 type promoter is degenerate (Hengge-Aronis 2002), the putative − 35 sequence of PectA2 (GTCTCG) is also not well conserved. With high frequency, RACE-PCR also mapped a transcription start point of C in the upstream ectB (Fig. 2b). However, no conservative sequences were found in front of its start point, and thereby the promoter type needs to be further verified.
To confirm the osmotic regulation of ectAB promoters, E. coli strains carrying promoter fusion plasmids (pkk-Y2-PectA and pkk-Y2-PectB) were measured for CAT activity in the presence of various ionic and nonionic osmolytes. For comparison, glycerol, which cannot establish an osmotic gradient across the cytoplasmic membrane, was also tested (Czech et al. 2018). As shown in Fig. 3a, the transcriptional activity of the ectA promoter was significantly triggered by both ionic (NaCl and KCl) and nonionic (sucrose) solutes, while glycerol failed to stimulate ectA promoter activity. In addition, a consistent increase in CAT activity was observed in the medium with elevated NaCl concentrations; an approximately 53 % increase in CAT activity was detected in the presence of 0.5 M NaCl when compared to the activity without NaCl. In contrast to the osmotic response of the ectA promoter, the CAT activity of pkk-Y2-PectB was non-osmotically regulated, with identical activities were observed when stimulated by various ions (Fig. 3b).
Screening strong and constitutive promoter from H. hydrothermalis Y2
Based on the results that the ectA promoter is osmotically controlled and has lower transcriptional activity, we engaged to screen strong promoters from strain Y2 for promoter substitution of ectA. As shown in Supplementary Table 3, 15 genes with high expression levels in strain Y2 were selected from the transcriptional data (Cheng et al. 2016). Approximately 1000 bp sequences upstream of each candidate gene were firstly used for promoter activity evaluation. As shown in Fig. 4, the recombinant strain carrying plasmid pKK-Y2-2155 displayed a protruding high CAT activity, with 5200 CAT activity per µg of protein observed. This was 80 fold higher than that of the original chloramphenicol promoter in the control group and was 12 fold higher than that of the secondary activity possessed by pKK-Y2-1913. Genomic annotation revealed that the promoter fused to recombinant pKK-Y2-2155 is from an outer membrane pore protein E (Eporin), which is located on the outer membrane and acts as a channel for passive diffusion of nutrients and other solutes (Lugtenberg et al. 1978).
In the genome of H. hydrothermalis Y2, a 265 bp intergenic region is located upstream of the Eporin coding gene and shows 89 % identity with a constitutive porin promoter that had been identified in Halomonas sp. TD1 (Li et al. 2016). In the 265-bp sequence of Eporin, a 210-bp region matched well with this homolog promoter of strain TD1, with only 21 bp differences observed. Furthermore, the 40-bp core region as well as the − 10 (GTATAGAGT) and − 35 (TTGCGT) elements, were completely conserved between these two promoters (Fig. 5). We therefore truncated the initial length of 1000 bp to the intergenic region (265 bp) and compared its expression with that of recombinant pKK-Y2-2155. As a result, minor differences in CAT activity was detected (Fig. 4), indicating that this 265-bp region is an intact promoter for gene transcription.
Schematic diagram of porin promoter and its flanking genes that identified in transcriptome analysis. The underlined sequence is the 210-bp promoter region. The RBS sequence are written in bold. The red region is the core region identical to the porin promoter in Halomonas sp. TD1, in which the − 10 and − 35 sequences are written in bold
Ectoine synthesis by the promoter replacement strain
To evaluate the promoter activity in ectoine production, the entire region upstream of ectA (258 bp) was substituted with the 265-bp porin promoter (P265) in the constructed strain p/Y2/△ectD/△doeA. In 500-ml baffle-bottomed flasks with various NaCl concentrations, the fusion strain exhibited growth profiles similar to those of Y2/△ectD/△doeA (Fig. 6a, c). In compared with the ectoine productivities of Y2/△ectD/△doeA (Fig. 6b), the productivities of p/Y2/△ectD/△doeA were increased at all tested salinities (Fig. 6d). Strikingly, strain p/Y2/△ectD/△doeA displayed a much higher synthetic ability in the low-salinity medium, along with a higher excretion ratio. For example, totally 1.22 g ectoine l−1 was synthesized by the p/Y2/△ectD/△doeA strain, while ectoine production of Y2/△ectD/△doeA was only 0.65 g l−1. Obviously, additional ectoine of strain p/Y2/△ectD/△doeA was excreted into the medium (Fig. 6d). This is in agreement with the linear relationship between the intracellular compatible solute content and salt concentration of the medium (Kuhlmann and Bremer 2002; Kunte 2006), i.e., these two strains retained similar concentrations of ectoine in cells under the same saline stress (Blue columns in Fig. 6b, d). Differently, the promoter replacement strain could synthesize more ectoine, and thus more products were excreted into a lower salinity media (Yellow columns, 20, 40, 60 g NaCl l−1). In contrast, as the saline stress outside increased to 80 g l−1, more compatible solutes were required for osmoregulation and no additional ectoine could be excreted.
The ectoine-producing ability of p/Y2/△ectD/△doeA was further investigated in a 1-l fermenter with an initial 20 g NaCl l−1 and final 60 g NaCl l−1. During the 48 h fermentation period, the p/Y2/△ectD/△doeA strain underwent rapid growth and reached to the maximum biomass (26.8 g CDW l−1) after 18 h of incubation, after which the biomass decreased (Fig. 7). In contrast, the ectoine titer increased constantly and achieved a 11.5 g ectoine l−1 after 36-h fermentation, with a 0.32 g ectoine l−1 h−1 and a specific productivity of 512.2 mg (g DCW)−1 (Fig. 7). The total consumed MSG was 42.1 g l−1, and the MSG yield was 0.27 g ectoine g−1.
Discussion
In most ectoine-producing Halomonas strains, the synthesis of ectoine is strictly salt dependent. Based on RACE-PCR results of H. hydrothermalis Y2, the putative promoters upstream of ectA are assumed to be the σ70 and σ38 type. In E. coli, the σ38 type promoter with the so-called G-element was previously suggested as the osmotic promoter that is responsible for salinity responses during ectoine synthesis (Lee and Gralla 2004). However, recent studies in C. salexigens DSM 3043T and H. elongata 2581T, and even in E. coli, demonstrated that σ38 is not essential for osmotic regulation of ectoine gene transcription (Typas et al. 2007; Salvador et al. 2015; Stiller et al. 2018). In contrast, the σ70 promoter region was demonstrated to be sufficient to establish a salt-related response in the ectoine gene cluster of P. stutzeri A1501 (Czech et al. 2018).
In this study, based on the findings that the upstream sequences of ectA contain both σ70 and σ38 promoters, the whole region upstream of ectA in a previously constructed Y2/△ectD/△doeA mutant was replaced by a constitutive promoter P265 that possesses high transcriptional activity. This promoter constitutively regulates the transcriptional expression of an outer membrane porin protein and is also identified in some moderate halophiles (Li et al. 2016; Nagayoshi et al. 2006). As expected, the resulting strain p/Y2/△ectD/△doeA showed a decreased salt dependence in ectoine synthesis. In addition, attribute to the strong transcriptional activity of promoter P265, the ectoine productivity increased in all tested saline media. The most striking difference was observed in the low-salinity media, in which more ectoine was synthesized and excreted into the medium. We suspected that p/Y2/△ectD/△doeA produced more ectoine than the cell requirement for balancing the osmolality, and therefore excessive ectoine can be continuously excreted into the medium. In the 1-l auto-controlled fermenter containing 60 g NaCl l−1, the promoter fusion strain also displayed a robust ability to synthesize ectoine, with a maximum titer of 11.5 g ectiube l−1 after 36 h of fed-batch fermentation and high excretion ratio of 67 %. The ectoine productivity was 0.32 g l−1 h−1, which was higher than our previously reported 10.5 g ectoine l−1 after 48 h of fermentation (0.22 g l−1 h−1), by strain Y2/△ectD/△doeA/△mrp that was cultured under the same conditions (Zhao et al. 2019).
It is worth noting that partial salt dependence was also observed during the ectoine synthesis of strain p/Y2/△ectD/△doeA. This is in accordance with the previous observation in P. stutzeri strain, where a series of mutants were constructed by altering the σ70 promoter region upstream of ectA. As a result, the basal ectA promoter activity was significantly enhanced in the absence of osmotic stress, while an increased promoter activity was maintained at high salinity. It was therefore proposed that osmotic regulation of ect gene expression is not determined merely by the DNA sequence that resides in the canonical promoter elements (Czech et al. 2018).
References
Becker J, Schäfer R, Kohlstedt M, Harder BJ, Borchert NS, Stöveken N, Wittmann C (2013) Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb Cell Fact 12:110
Calderon MI, Vargas C, Rojo F, Iglesias-Guerra F, Csonka LN, Ventosa A, Nieto JJ (2004) Complex regulation of the synthesis of the compatible solute ectoine in the halophilic bacterium Chromohalobacter salexigens DSM 3043T. Microbiology 150:3051–3063
Cheng B, Meng Y, Cui Y, Li C, Tao F, Yin H, Yang C, Xu P (2016) Alkaline response of a halotolerant alkaliphilic Halomonas strain and functional diversity of its Na+(K+)/H+ antiporters. J Biol Chem 91:26056–26065
Czech L, Poehl S, Hub P, Stoeveken N, Bremer E (2018) Tinkering with osmotically controlled transcription allows enhanced production and excretion of ectoine and hydroxyectoine from a microbial cell factory. Appl Environ Microbiol 84:01772–01717
Hengge-Aronis R (2002) Stationary phase gene regulation: what makes an Escherichia coli promoter sigma S-selective? Curr Opin Microbiol 5:591–595
Jadhav K, Kushwah B, Jadhav I (2018) Insight into compatible solutes from halophiles: exploring significant applications in biotechnology. Microbial Bioprospecting for Sustainable Development, pp 978–981
Kuhlmann AU, Bremer E (2002) Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl Environ Microbiol 68:772–783
Kunte HJ (2006) Osmoregulation in bacteria: compatible solute accumulation and osmosensing. Environ Chem 3:94–100
Lee SJ, Gralla JD (2004) Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol Cell 14:153–162
Li T, Li T, Ji W, Wang Q, Zhang H, Chen GQ, Lou C, Qi OY (2016) Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp. Biotech J 11:219–227
Lugtenberg B, Boxtel RV, Verhoef C, Alphen WV (1978) Pore protein of the outer membrane of Escherichia coli K12. FEBS Lett 96:0–105
Nagayoshi C, Tokunaga H, Hayashi A, Harazono H, Hamasaki K, Ando A, Tokunaga M (2006) Efficient expression of haloarchaeal nucleoside diphosphate kinase via strong porin promoter in moderately halophilic bacteria. Protein Pept Lett 13:611–615
Oren A (2002) Halophilic microorganisms and their environments. Kluwer Academic Publishers, (Eds.). Dordrecht
Oren A (2018) Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Systems 4:1–13
Pejin B, Ciric A, Glamoclija J, Nikoli M, Sokovic M (2015) In vitro anti-quorum sensing activity of phytol. Nat Prod Res 29:374–377
Salvador M, Argandoña M, Pastor JM, Bernal V, Cánovas M, Csonka LN, Nieto JJ, Vargas C (2015) Contribution of RpoS to metabolic efficiency and ectoines synthesis during the osmo- and heat‐stress response in the halophilic bacterium Chromohalobacter salexigens. Env Microbiol Rep 7:301–311
Schwibbert K, Marin-Sanguino A, Bagyan I, Heidrich G, Lentzen G, Seitz H, Rampp M, Schuster SC, Klenk HP, Pfeiffer F, Oesterhelt D, Kunte HJ (2011) A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581T. Environ Microbiol 13:1973–1994
Seip B, Galinski EA, Kurz M (2011) Natural and engineered hydroxyectoine production based on the Pseudomonas stutzeri ectABCD-ask gene cluster. Appl Environ Microbiol 7:1368–1374
Stiller LM, Galinski EA, Emhj W (2018) Engineering the salt-inducible ectoine promoter region of Halomonas elongata for protein expression in a unique stabilizing environment. Genes 9:184
Typas A, Stella S, Johnson RC, Hengge R (2007) The σ35 sequence location and the Fis-sigma factor interface determine σS selectivity of the proP (P2) promoter in Escherichia coli. Mol Microbiol 63:780–796
Wösten MMSM (1998) Eubacterial sigma-factors. FEMS Microbiol Rev 22:127–150
Yang C, Wang Z, Li Y, Niu Y, Du M, He X, Ma C, Tang H, Xu P (2010) Metabolic versatility of halotolerant and alkaliphilic strains of Halomonas isolated from alkaline black liquor. Bioresour Technol 101:6778–6784
Zhao Q, Li S, Lv P, Sun S, Ma C, Xu P, Su H, Yang C (2019) High ectoine production by an engineered Halomonas hydrothermalis Y2 in a decreased salinity medium. Microb Cell Fact 18:18412
Funding
This work was funded by the National Natural Science Foundations of China (Grant nos. 31870094 and 31670109).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. LS, SY and ZQ performed the experiments. YC and LS design the research. LS, ZQ, WW and DX contribute to data analysis. YC, SY, and LY wrote and revise the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, S., Shang, Y., Zhao, Q. et al. Promoter engineering for high ectoine production in a lower saline medium by Halomonas hydrothermalis Y2. Biotechnol Lett 43, 825–834 (2021). https://doi.org/10.1007/s10529-021-03084-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-021-03084-3