Abstract
Objective
To identify useful native promoters of Corynebacterium glutamicum for fine-tuning of gene expression in metabolic engineering.
Results
Sixteen native promoters of C. glutamicum were characterized. These promoters covered a strength range of 31-fold with small increments and exhibited relatively stable activity during the whole growth phase using β-galactosidase as the reporter. The mRNA level and enzymatic activity of the lacZ reporter gene exhibited high correlation (R 2 = 0.96) under the control of these promoters. Sequence analysis found that strong promoters had high similarity of the -10 hexamer to the consensus sequence and preference of the AT-rich UP element upstream the -35 region. To test the utility of the promoter library, the characterized native promoters were applied to modulate the sucCD-encoded succinyl-CoA synthetase expression for l-lysine overproduction.
Conclusions
The native promoters with various strengths realize the efficient and precise regulation of gene expression in metabolic engineering of C. glutamicum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynebacterium glutamicum, a non-pathogenic, non-sporulating Gram-positive bacterium is generally recognized as a safe (GRAS) organism for industrial production of bio-based chemicals such as amino acids, nucleotides and so on (Becker and Wittmann 2015). Genetic tools including various plasmids, promoters and other regulatory elements are indispensable to study the physiological characteristics and improve the performance of industrial strains (Nešvera and Pátek 2011; Zhao et al. 2016). A few well-defined inducible promoters, such as P tac and P araBAD from Escherichia coli, have been widely used to regulate gene expression in C. glutamicum (Zhang et al. 2012). However, they could not exhibit the optimal activities due to the different mechanisms of regulations and the inefficient permeability of the inducer (Yim et al. 2013). Besides, two synthetic promoter libraries have been constructed by random mutation to modulate gene expression (Rytter et al. 2014; Yim et al. 2013).
Compared to synthetic promoters, native promoters are easily accessed and more efficient in the metabolic engineering of C. glutamicum. Two strong promoters of the sod gene encoding superoxide dismutase and the tuf gene encoding translational elongation factor have been widely used to enhance the expression of key genes for the overproduction of l-lysine (Becker et al. 2011). Besides, mutants of the well-characterized native promoter P dapA had been applied to modulate citrate synthase flux in metabolic engineering for l-lysine production (Ooyen et al. 2012). Despite the identification and analysis of many native promoter sequences in C. glutamicum (Pátek and Nešvera 2011), the unknown transcription activities of these promoters makes them unavailable for metabolic engineering.
In this study, 16 native promoters of C. glutamicum were characterized definitely. These promoters exhibited wide-ranged strengths and relatively stable activity during the whole growth phase, which facilitated the fine-tuning of gene expression. The sequence characteristics of these promoters were found to be relevant to their strengths. To examine the potential of the native promoters, two low-strength promoters were used to modulate the expression of the sucCD gene, which successfully increased l-lysine production.
Materials and methods
Strains, plasmids and growth conditions.
Bacterial strains and plasmids used in this study are listed in Supplementary Table 1. Escherichia coli EC135 strain was used for gene cloning. C. glutamicum and E. coli strains were routinely grown in lysogeny broth (LB) at 30 and 37 °C, respectively. When needed, 20 µg chloramphenicol ml−1 for E. coli and 10 µg chloramphenicol ml−1 for C. glutamicum was added; 50 µg kanamycin ml−1 for E. coli and 25 µg kanamycin ml−1 for C. glutamicum was added to the medium. For l-lysine production, pre-cultures were grown for 8 h in the complex medium containing (per liter): 20 g glucose, 5 g (NH4)2SO4, 2 g urea, 5 g yeast extract, 5 g tryptone, 1.6 g K2HPO4, 0.4 g MgSO4·7H2O, 10 mg FeSO4·7H2O, 10 mg MnSO4·H2O, 1 mg ZnSO4·7H2O, 0.1 mg CuSO4·5H2O, 0.4 mg thiamine. HCl, and 50 μg biotin and then inoculated into 30 ml fermentation medium containing (per liter): 40 g glucose, 2 g yeast extract, 15 g (NH4)2SO4, 2.5 g KH2PO4, 0.4 g MgSO4·7H2O, 10 mg FeSO4·7H2O, 10 mg MnSO4·H2O, 1 mg ZnSO4·7H2O, 0.1 mg CuSO4·5H2O, 200 μg thiamine.HCl, and 100 μg biotin in 500 ml baffled shake-flasks to a final OD600 of 1. Fermentation was conducted for 48 h. During the fermentation, glucose was fed from a 600 g glucose l−1 stock solution when the residual glucose concentration was below 10 g glucose l−1 and NH4OH was added to maintain the pH at 7.
Construction of plasmids and strains
The genomic DNA, PCR product and plasmid DNA was isolated using the genomic isolation kit, gel extraction kit and plasmid isolation kit, respectively (Tiangen, Beijing, China). All the primers used in this study are listed in Supplementary Table 2. The native promoters of C. glutamicum were amplified using the genome DNA of C. glutamicum ATCC 13032 as the template. The entire open reading frame of the lacZ (β-galactosidase) gene was amplified from E. coli W3110, ligated to the pXMJ19 plasmid in which the tac promoter and repressor regions were removed by restriction enzymes, and thus the promoter probe vectors were obtained. Different promoters were ligated into the promoter probe vectors and transformed into C. glutamicum by electroporation. All modifications introduced into the C. glutamicum chromosome were conducted through the homologous recombination plasmid pK18mobsacB. For gene overexpression in the lysine-producing strains, the lysC C932T (feedback resistant aspartokinase), ddh (diaminopimelate dehydrogenase) and lysA (diaminopimelate decarboxylase) were amplified and ligated into the pXMJ19 by Gibson assembly in an expression cassette under the control of the isopropy-β-d-thiogalactoside (IPTG) inducible tac promoter (Becker et al. 2011). The consensus RBS sequence ‘AAAGGAGGA’ was inserted in the front of the open reading frame of lysC C932T, ddh and lysA to increase their expression. The recombinant plasmid was then transformed into the basic lysine-producing strain and the resulted strain LYS-1 was employed to evaluate the effect of promoter engineering. All the modifications were verified by PCR amplification and DNA sequencing.
β-Galactosidase assay
The fluorogenic substrate, 4-methylumbelliferyl-d-galactopyranoside (MUG), was used to measure β-galactosidase activity. C. glutamicum cells were grown overnight in LB and 40 μl of the cultures were transferred to 96-well microplates containing 80 μl Z-buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0) and the corresponding cell density was evaluated by determining the OD595. 20 μl MUG dissolved in dimethyl sulfoxide were added to each well, and the samples were incubated at room temperature for 15 min. The reaction was stopped with 1 M Na2CO3. The degree of fluorescence generated by β-gal-dependent MUG hydrolysis was quantitated in a microplate fluorometer (360 nm excitation filter, 460 nm emission filter). The β-gal activities were calculated as MUG Units as reported (Vidal-Aroca et al. 2006). When needed, 0.5 mM IPTG was added to induce the lacZ gene expression.
Quantitative reverse transcriptional PCR (qRT-PCR)
Total RNA was extracted using the RNAprep Pure Cell/Bacteria Kit (TIANGEN, Beijing, China) and then stored at − 80 °C until use. The reverse transcription was performed with the specific primers of the target gene using the FastQuant RT Kit (Tiangen, Beijing, China). Quantitative PCR was performed with BRYT Green from the GoTaq qPCR master mix (Promega, USA) by the LightCycler 96 Real-Time PCR System (Roche, Switzerland). The rpoB gene of C. glutamicum was used as the reference gene. Negative controls were done to exclude the DNA and other contaminations. The specificity of the qPCR products was verified by a melting curve analysis. Data analysis was facilitated using the LightCycler 96 software.
Analytic procedures
The cell growth was monitored from the OD600 value and the glucose concentration was detected by an SBA-40D biosensor automatic analyzer (Institute of Biology of Shandong Province Academy of Sciences, Shandong, China). l-Lysine concentration in the supernatant of the fermentation culture was determined by HPLC with a Zorbax Eclipse-AAA column (4.6 mm × 150 mm, 5 μm; Agilent Technology, USA) at 40 °C after automated online derivatization using o-phthalaldehyde (OPA). The elution was performed using a gradient of reagent A (40 mM NaH2PO4·2H2O, pH 7.8) and reagent B (methanol/acetonitrile/water = 45:45:10, by vol.) at 2 ml/min. Elution gradients were: 0 to 1.9 min, 0% B; thenh to 18.1 min, 57% B; to 18.6 min, 100% B; 22.3 min, 100% B; 23.2 min, 0% B; 26 min, 0% B. The eluate was monitored at 338 nm.
Flux balance analysis
A reconstructed genome-scale metabolic model of C. glutamicum ATCC 13032 named iCW773, which could accurately predict the flux distributions of cells cultured under different growth conditions, was used for in sillico simulation based on Cobra Toolbox 2.05 and MatLab 2014a with LP solver GLPK (Zhang et al. 2017). For flux balance analysis (FBA), the phenotypes were calculated for a fixed glucose uptake rate of 4 mmol glucose g DCW−1 h−1 using growth and lysine yield as a cellular objective function, respectively. To investigate the effect of succinyl-CoA synthetase (SUCOAS) flux on lysine yield and cell growth, relative SUCOAS fluxes were used to perform the robustness analysis by FBA.
Results
Evaluation of the native promoter strengths in C. glutamicum
To expand the tools for fine-tuning of gene expression, promoters of C. glutamicum chromosomal genes involved in the central carbon and amino acid metabolism were screened to analyze their strengths. The frequently used native promoter P tuf and P sod were also conducted for activity evaluation and the hybrid promoter P tac was applied as the control. The 200 bp sequences upstream of the translation start sites of the sixteen selected genes, which contained the full length promoters and the native 5′-untranslated regions (5′-UTRs), were amplified and ligated into a promoter probe vector with lacZ as the reporter gene. The strengths of these promoters were evaluated by determining the specific activities of β-galactosidase in C. glutamicum.
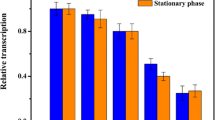
To test the stability of the promoters, cells were cultivated and harvested at 6, 12 and 24 h, which correspond to the mid-growth phase, the late-growth phase and the stationary phase, respectively (Fig. 1a, b). As shown in the determination of the β-galactosidase activity, a wide range of promoter activities was covered from 73 to 2257 MUG units, corresponding to a span of 31-fold of the promoter activity. Most of the promoters exhibited relatively stable activity throughout the cultivation, i.e., P pgk and other promoters in the H-group yielded relatively high β-galactosidase activity in different growth phases, whereas promoters in the L-group only generated low β-galactosidase activity (Fig. 1c).
Promoter strength analysis as presented by the β-galactosidase activities of the lacZ reporter. a, b Growth of C. glutamicum strains containing different promoter-probe vectors. c Cells were collected at 6, 12 and 24 h for β-galactosidase activity assay. The P tac was used as a control. The error bars indicate the standard deviations of biological triplicate
Transcriptional analysis of the promoters
To accurately evaluate the activity of these promoters, the relative mRNA levels of the lacZ under control of different promoters were analyzed by qRT-PCR and normalized to that under the control of P tac . The mRNA levels of the lacZ gene spanned a range of 33-fold and the orders of most promoter strengths ranked by the transcription level were in accordance with those ranked by the β-galactosidase activity (Fig. 2a). The correlation matrix between the β-galactosidase activity and the transcription level showed a linear correlation (R 2 = 0.96) (Fig. 2b), which indicated that the reporter gene could be precisely modulated by the representative promoters at the transcription level.
Sequence analysis of the C. glutamicum native promoters
As all of the transcription start sites (TSS) and core region of the selected promoters are available (Mentz et al. 2013; Pátek and Nešvera 2011), the sequences upstream the TSS including the -10 and -35 region were aligned. The conserved hexamer TANANT was found in the -10 region whereas sequences in the -35 region were much less conserved in the sixteen promoters (Crooks et al. 2004; Pátek and Nešvera 2011). Besides, the dinucleotide up- and downstream of the -10 region hexamer was relatively less conserved in the sixteen promoters (Supplemented Fig. 1). To investigate the relationship between the sequence feature and the strength of H-group and L-group promoters, the core elements of these promoters were aligned using the WebLogo software to compare the abundance of each nucleotide at different positions of the promoters. The -10 core region of the H-group promoters exhibited high similarity to the most conserved TATAAT hexamer of the C. glutamicum promoters. The conserved TTG in the -35 region showed higher abundance in the H-group promoters compared to the L-group promoters. Besides, the AT-rich UP element located in the upstream of the -35 region showed preference in the H-group promoters (Fig. 3).
Sequence logo of the 9 strong promoters and the 6 weak promoters using the Weblogo software. a, b Sequence logo of the -10 region of the strong promoters and the weak promoters, respectively. c, d Sequence logo of the -35 region of the strong promoters and the weak promoters, respectively. The -10 and -35 regions were marked by a grey box and the UP element upstream the -35 region was marked by a dashed box
Enhanced l-lysine production by promoter activity modulation
To evaluate the application potential of native promoters in metabolic engineering, the promoters were used to modulate the gene expressions for l-lysine overproduction. A basic lysine-producing strain derived from C. glutamicum ATCC 13032 was constructed based on the metabolic engineering strategies described in the literature (Becker et al. 2011), which contained mutated hom T176C (homoserine dehydrogenase) and pyc C1372T (pyruvate carboxylase) in the chromosome, pck (PEP carboxykinase) deletion and the additional copy of the mutated pyc C1372T and dapB (dihydrodipicolinate reductase) in the pck locus. Besides, lysC C932T, ddh and lysA were overexpressed on the plasmid pXMJ19 and the resulted strain LYS-1 served as the platform to evaluate the effect of promoter engineering.
In addition to carbon building blocks, lysine biosynthesis through the succinylase branch further involves succinyl-CoA as the activator of the pathway intermediates for introducing nitrogen, which made sucCD-encoded SUCOAS as a promising target for metabolic engineering (Kind et al. 2013). We used a recently reconstructed genome-scale metabolic model of C. glutamicum ATCC 13032 (iCW773) for in silico simulation to evaluate the impact of SUCOAS flux on growth and lysine production (Zhang et al. 2017). The simulation results showed that decreased SUCOAS flux would increase lysine yield and maintain the optimal cell growth at the same time (Fig. 4a). For sucCD downregulation, weak promoters P hom and P dapA were delivered into the region upstream of the sucCD operon in strain LYS-1, replacing its native promoter and inherent regulation modality. The mRNA level of sucC decreased by 47 and 64% in the corresponding strains LYS-2 and LYS-3, respectively (Fig. 4b). As down-regulating sucCD, LYS-2 and LYS-3 exhibited similar growth compared to the platform strain LYS-1, whereas the lysine production increased by 16 and 20% in strains LYS-2 and LYS-3, respectively (Fig. 4c).
Modulation of sucCD by the native promoters and the effect on l-lysine biosynthesis. a Simulation of the impact of SUCOAS flux on lysine production for robustness analysis. b The transcription levels of sucC as controlled by the promoter P hom (LYS-2) and P dapA (LYS-3). c Effect of the down-regulation of sucCD on the lysine production. The LYS-1 strain was applied as the control. The error bars indicate the standard deviations of biological triplicate. * and ** indicates P < 0.05 and P < 0.01, respectively
Discussion
Sixteen efficient native promoters with wide-ranged strengths were characterized for precise gene modulation in C. glutamicum. Besides the native strong promoter P tuf and P sod , the promoter of the pgk gene in the glycolytic pathway was demonstrated to be an extremely strong promoter, which was the first native promoter found to exhibit higher activity than the P tac . In addition, weak promoters such as P dapA , P dapB , and P ddh were presented, which could be applied in down-regulating gene expression instead of gene deletion. Artificial promoters by fusing core sequences of native promoters and different operator regions have been developed to implement controllable gene expression (Park et al. 2008). The H-group promoters in this study could also be the candidates to design the synthetic promoter by fusing with specific sequence recognized by regulatory protein for the inducible expression of target genes. Additionally, these promoters with different strengths would be promising elements for fine-tuning gene expression on the expression vector in C. glutamicum as previous reported (Lee 2014).
Sequence characteristics of C. glutamicum promoters were analyzed on the basis of the available transcriptomic data obtained by high-throughput RNA-sequencing (Albersmeier et al. 2017; Mentz et al. 2013). Being indispensable for sigma factor of the RNA polymerase holoenzyme binding, -10 region was the dominating element in promoter structure. The most conserved consensus sequence TATAAT existed in the -10 region of the H-group promoters as reported previously (Pátek et al. 2003; Pátek and Nešvera 2011), indicating the essentiality of -10 region in the strength of promoters. For most of the cases, promoter strength depends mainly on the similarity to the consensus sequence (Albersmeier et al. 2017). Despite the extremely high strength of P pgk promoter, the -10 region of P pgk promoter showed a low similarity to the consensus hexamer, which was similar as other promoters whose sequence also did not follow the conservation rule. It might be attributed to that the strength of P pgk promoter was evolved to increase the fitness of the cell due to the important role of the pgk gene in the glycolysis pathway. Besides, UP element (A + T rich) which interacted with C-terminal domain of the RNA polymerase α-subunit to enhance the transcription activity was found in the strong promoters (Pátek and Nešvera 2011).
Being σA-dependent, these promoters exhibited stable activity during the whole growth phase, however, the σA-dependent house-keeping promoters could also be regulated (Han et al. 2007). In our results, the strength of P glyA increased to a higher level at the stationary phase, which attributed to the up-regulation of glyA expression by the activator GlyR in the stationary phase (Schweitzer et al. 2009). Additionally, the constitutively expressed glycolytic genes of C. glutamicum are induced in the presence of glucose (Han et al. 2007). Besides, the gnd gene encoding the 6-phosphogluconate dehydrogenase was proved to be induced in the presence of gluconate by the transcriptional regulators GntR1 and RamA (Tanaka et al. 2012). Therefore, we inferred that P pgk , P fba and P pyc as well as the P gnd would be induced to exhibit higher activities in the presence of inducers (glucose or gluconate) when compare to those presented in the commonly used LB medium.
In conclusion, the promoters with various strengths characterized in our study will facilitate the precise modulation of gene expression in metabolic engineering of C. glutamicum.
References
Albersmeier A, Pfeifer-Sancar K, Rückert C, Kalinowski J (2017) Genome-wide determination of transcription start sites reveals new insights into promoter structures in the actinomycete Corynebacterium glutamicum. J Biotechnol 257:99–109
Becker J, Wittmann C (2015) Advanced biotechnology: metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew Chem Int Ed 54:3328–3350
Becker J, Zelder O, Häfner S, Schröder H, Wittmann C (2011) From zero to hero—Design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng 13:159–168
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Han SO, Inui M, Yukawa H (2007) Expression of Corynebacterium glutamicum glycolytic genes varies with carbon source and growth phase. Microbiology 153:2190–2202
Kind S, Becker J, Wittmann C (2013) Increased lysine production by flux coupling of the tricarboxylic acid cycle and the lysine biosynthetic pathway—Metabolic engineering of the availability of succinyl-CoA in Corynebacterium glutamicum. Metab Eng 15:184–195
Lee J (2014) Development and characterization of expression vectors for Corynebacterium glutamicum. J Microbiol Biotechnol 24:70–79
Liu Q, Ouyang SP, Kim J, Chen GQ (2007) The impact of PHB accumulation on l-glutamate production by recombinant Corynebacterium glutamicum. J Biotechnol 132:273–279
Mentz A, Neshat A, Pfeifer-Sancar K, Pühler A, Rückert C, Kalinowski J (2013) Comprehensive discovery and characterization of small RNAs in Corynebacterium glutamicum ATCC 13032. BMC Genom 14:714
Nešvera J, Pátek M (2011) Tools for genetic manipulations in Corynebacterium glutamicum and their applications. Appl Microbiol Biotechnol 90:1641–1654
Ooyen JV, Noack S, Bott M, Reth A, Eggeling L (2012) Improved l-lysine production with Corynebacterium glutamicum and systemic insight into citrate synthase flux and activity. Biotechnol Bioeng 109:2070–2081
Park JU, Jo JH, Kim YJ, Chung SS, Lee JH, Lee HH (2008) Construction of heat-inducible expression vector of Corynebacterium glutamicum and C. ammoniagenes: fusion of lambda operator with promoters isolated from C. ammoniagenes. J Microbiol Biotechnol 18:639–647
Pátek M, Nešvera J (2011) Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol 154:101–113
Pátek M, Nešvera J, Guyonvarch A, Reyes O (2003) Promoters of Corynebacterium glutamicum. J Biotechnol 104:311–323
Rytter JV, Helmark S, Chen J, Lezyk MJ, Solem C, Jensen PR (2014) Synthetic promoter libraries for Corynebacterium glutamicum. Appl Microbiol Biotechnol 98:2617–2623
Schweitzer JE, Stolz M, Diesveld R, Etterich H, Eggeling L (2009) The serine hydroxymethyltransferase gene glyA in Corynebacterium glutamicum is controlled by GlyR. J Biotechnol 139:214–221
Tanaka Y, Ehira S, Teramoto H, Inui M, Yukawa H (2012) Coordinated regulation of gnd, which encodes 6-phosphogluconate dehydrogenase, by the two transcriptional regulators GntR1 and RamA in Corynebacterium glutamicum. J Bacteriol 194:6527–6536
Vidal-Aroca F, Giannattasio M, Brunelli E, Vezzoli A, Plevani P, Muzi-Falconi M, Bertoni G (2006) One-step high-throughput assay for quantitative detection of β-galactosidase activity in intact gram-negative bacteria, yeast, and mammalian cells. Biotechniques 40:433–440
Yim SS, An SJ, Kang M, Lee J, Jeong KJ (2013) Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol Bioeng 110:2959–2971
Zhang Y, Shang XL, Lai SJ, Zhang GQ, Liang Y, Wen TY (2012) Development and application of an arabinose-inducible expression system by facilitating inducer uptake in Corynebacterium glutamicum. Appl Environ Microbiol 78:5831–5838
Zhang Y, Cai JY, Shang XL, Wang B, Liu SW, Chai X, Tan TW, Zhang Y, Wen TY (2017) A new genome-scale metabolic model of Corynebacterium glutamicum and its application. Biotechnol Biofuels 10:169
Zhao ZH, Liu XX, Zhang W, Yang YK, Dai XF, Bai ZH (2016) Construction of genetic parts from the Corynebacterium glutamicum genome with high expression activities. Biotechnol Lett 38:2119–2126
Acknowledgements
This work was supported by grants from National Hi-Tech Research and Development Program of China (2014AA021203), the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (KFJ-EW-STS-078) and National Natural Science Foundation of China (31100074).
Supporting information
Supplementary Table 1—Strains and plasmids used.
Supplementary Table 2—Primers used.
Supplementary Fig. 1—Sequence analysis of the sixteen promoters.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal participants
The research performed did not involve human participants and/or animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shang, X., Chai, X., Lu, X. et al. Native promoters of Corynebacterium glutamicum and its application in l-lysine production. Biotechnol Lett 40, 383–391 (2018). https://doi.org/10.1007/s10529-017-2479-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-017-2479-y