Abstract
Objectives
To elucidate the mechanism of cellulase signal transduction in filamentous fungi including the components of the cellulase induction pathway.
Results
Neurospora crassa ncw-1 encodes a non-anchored cell wall protein. The absence of ncw-1 increased cellulase gene expression and this is not due to relieving carbon catabolite repression mediated by the cre-1 pathway. A mutant lacking genes encoding both three major β-glucosidase enzymes and NCW-1 (Δ3βGΔncw-1) was constructed. Transcriptome analysis of the quadruple mutant demonstrated enhanced expression of cellodextrin transporters after ncw-1 deletion, indicating that ncw-1 affects cellulase expression and production by inhibiting the uptake of the cellodextrin.
Conclusions
NCW-1 is a novel component that plays a critical role in the cellulase induction signaling pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Filamentous fungi secrete a wide variety of lignocellulolytic enzymes that convert complex biomass into fermentable sugars. This capacity has been exploited by industry to produce biofuels and biochemicals (Sun and Cheng 2002). However, the high cost of lignocellulolytic enzyme production is a major bottleneck in the utilization of lignocellulosic biomass. A premise of the current work is that a better understanding of cellulase induction pathway in filamentous fungi could provide novel strategies to increase cellulase yield.
The filamentous fungus, Neurospora crassa, a native degrader of lignocellulose, has been investigated as a novel model for cellulase induction and synthesis. Using this system, β-glucosidases have been shown to play critical roles in cellulase induction. Deletion of three major β-glucosidases led to rapid and efficient induction of cellulase on cellobiose (Znameroski et al. 2012). In addition, two cellodextrin transporters, CDT-1 and CDT-2, contribute to both cellobiose uptake and cellulase inducer sensing (Znameroski et al. 2014). Although β-glucosidase and the cellodextrin transporter have been shown to be key components of the cellulase induction pathway (Zhang et al. 2013; Zhou et al. 2012), little is known about how this pathway is regulated and what other components might participate in this pathway.
The fungal cell wall is the interphase with the surrounding environment and plays crucial roles in signal sensing and transduction. N. crassa NCU05137 encodes a non-anchored cell wall protein (NCW-1). Deletion of this gene (or its homolog PDE01641) led to enhanced cellulase production in N. crassa (Tian et al. 2009) and Penicillium decumbens (Zhang et al. 2012). However, the mechanism by which deletion of ncw-1 enhanced cellulase production is unclear. To decipher this mechanism, we further investigated the critical roles of NCW-1 during cellulase induction. Here, we demonstrate that the improvement of cellulase production in the Δncw-1 is not a consequence of relief from carbon catabolite repression, and provide evidence that NCW-1 is indeed involved in cellulase induction by influencing cellodextrin transporter expression.
Materials and methods
Strains and culture conditions
Neurospora crassa wild-type (WT) strain and all single-gene deletion strains were obtained from the Fungal Genetics Stock Center. N. crassa strains were grown on Vogel’s minimal media (MM) (Tian et al. 2009). For liquid cultures, 107 conidia were inoculated into 100 ml Vogel’s salts with 2% (w/v) Avicel or 2% (w/v) sucrose in a 250 ml Erlenmeyer flask and cultivated at 25 °C in constant light and shaking (200 rpm).
Complementation of Δncw-1 and subcellular localization of NCW-1-GFP in N. crassa
WT genomic DNA was extracted as described previously (Lin et al. 2011). To complement Δncw-1, the ORF of ncw-1 was cloned by PCR with primers ORF-F and ORF-R, and then the fragment was inserted into the XbaI and PacI sites of pMF272 to form pMF272-Pc-ncw-1-gfp. The promoter of ncw-1 was cloned with primers PF and PR, and inserted into the NotI and XbaI sites of pMF272-Pc-ncw-1-gfp to result in pMF272-Pn-ncw-1-gfp. Plasmid DNA (10 μg) was transformed into a (his-3; Δncw-1) strain as described (Wang et al. 2015). The resulting complemented strains were termed Pc-ncw-1 and Pn-ncw-1. Observations of NCW-1 localization were performed with a fluorescence microscope.
To compare the differences in cell surface structure between the WT and Δncw-1, fungal conidia were incubated in MM at 25 °C for 40 h. The harvested mycelia were fixed in 0.1 M phosphate buffer (pH 7.2) containing 2.5% (w/v) glutaraldehyde. The samples were rinsed in thrice-distilled PBS, and dehydrated with a series of ethanol solutions (30–100%, v/v). After which the samples were dried by critical point drying (Leica CPD300) and coated with platinum (Hitachi E-1045). Observations were conducted using a scanning electron microscope (Hitachi SU8010) operating at 3 kV.
Enzyme activity and cellobiose consumption assay
Total extracellular proteins were determined with the Bradford assay (Bio-Rad Protein Assay kit). The endoglucanase and endoxylanse activities were assayed using the azo-CMC and azo-xylan kits (Megazyme), respectively, according to manufacturer’s instructions. p-Nitrophenyl-d-cellobioside (pNPC) and p-nitrophenyl-β-d-glucopyranoside (pNPG) were used as the substrates for the measurement of exoglucanase and β-glucosidase activities as described previously (Wang et al. 2015). The amount of cellobiose remaining in the supernatant was determined by HPLC with Aminex HPX-87H column (Bio-Rad) (Znameroski et al. 2012). All assays were performed in triplicate.
Biomass assay and rheological measurement
Mycelia grown on 2% (w/v) Avicel for 7 days were harvested, dried, and weighed. The relative proportion of mycelia to residual Avicel was calculated by solubilizing fungal biomass in boiling acetic nitric reagent as described (Li et al. 2014). The residual mass was washed thoroughly, dried, and reweighed.
Rheological properties of fermentation broth were determined by using a Brookfield digital HBDV-III Ultra rheometer according to the manufacturer’s instructions. Viscosity was measured as the torque exerted by the mycelial sample on the rheometer impeller at 150 rpm. Torque was recorded after the reading had stabilized (45–60 s). All assays were performed in triplicate.
RNA sequencing and data analysis
For gene expression analysis, mycelia were pre-cultured for 20 h in MM and transferred into Vogel’s salts containing 1% (w/v) cellobiose for an additional 4 h. Total RNA from frozen samples was extracted and treated as described previously (Li et al. 2014). RNA-seq was performed on the Illumina HiSeq 2000 platform of Beijing Genomics Institute (BGI, Shenzhen, China). Filtered clean reads were mapped against predicted transcripts from the N. crassa OR74R genome (version 12) using Tophat (version 2.0.8b). The alignment results were stored in SAM format files for subsequent analysis. The counts of reads that uniquely mapped to only one gene were calculated for each gene by Htseq-count (http://www-huber.embl.de/users/anders/HTSeq) using SAM files and genome annotation as input. Abundance for each transcript was calculated using the reads per kilobase per million (RPKM). Genes with altered expression were performed by using R package NOISeq (version 2.6.0) (Q value ≥ 0.95 used as threshold). Profiling data are listed in Supplementary Table 1 and the sequence data produced in this study can be accessed (GEO: GSE73838).
Quantitative real time-PCR
Quantitative PCR was performed using a CFX96 real-time PCR detection system (Bio-Rad) with reagents from TOYOBO (One-step qPCR Kit). All assays were performed in triplicate with actin (NCU04173) as the endogenous standard, according to the manufacturer’s instruction. The primers used in this study are listed in Supplementary Table 2.
Results and discussion
Deletion of ncw-1 increases biomass accumulation and broth viscosity
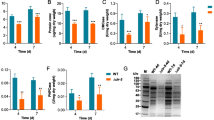
During 7 days growth on 2% (w/v) Avicel medium the broth of the ncw-1 mutant became more viscous than that of WT strain. The viscosity of Δncw-1 was significantly increased by twofold compared to that of WT strain (Fig. 1a). It is well known that both mycelial morphology and biomass concentration have significant influences on the rheological properties of a fermentation broth. Scanning electron microscopy demonstrated that Δncw-1 exhibited much smoother surface than the WT (Supplementary Fig. 1). Nevertheless, the ncw-1 mutant still showed a clump type of morphology in submerged culture, which was indistinguishable from that of WT. So we speculated that the ncw-1 mutant might grow faster on Avicel. Figure 1b shows that the biomass of the Δncw-1 mutant was 47% higher than that of WT strain. Thus, the change in rheological properties is probably due in part to increased growth.
Phenotype of Δncw-1 grown on Avicel. Conidia from Δncw-1 and wild-type (WT) strains were separately inoculated into Avicel medium and batch cultured for 7 days. The culture viscosity (a) and biomass accumulation (b) were measured. Values represent the means of at least three replicates, error bars show standard deviation. c Microscopic observation of NCW-1 subcellular localization in N. crassa. Scale bar is 10 μm
Additionally, the complementation analysis indicated that complementing Δncw-1 with a genomic clone of ncw-1 produced a strain indistinguishable from the wild type in all the phenotypic assays performed in this study (data not shown).
NCW-1 mainly localizes to the cell wall and hyphal tip
In order to obtain more detailed information about subcellular localization, we constructed a C-terminal GFP-tagged NCW-1 in the Δncw-1 strain under the control of the ccg-1 promoter (Pc-ncw-1 strain). Fluorescence microscopy indicated that the NCW-1 was localized to the surface of spores and hyphae. Intriguingly, NCW-1-GFP also accumulated at the hyphal tip area (Fig. 1c), where protein secretion occurs (Wosten et al. 1991). Since the cell wall at the hyphal tip is mostly composed of primary wall material with a comparatively porous structure, we hypothesized that NCW-1 could rapidly pass through this region and diffuse into the medium, which could also explain why NCW-1 comprises as much as 1.5% of the supernatant protein by weight (Phillips et al. 2011).
The enhanced cellulase production in the Δncw-1 mutant is not due to decreased carbon catabolite repression
The zinc finger transcription factor cre-1 is a key regulator in carbon catabolite repression. Deletion of cre-1 or its homolog could lead to enhanced cellulase production (Sun and Glass 2011). Since the disruption of ncw-1 enhances expression of cellulase genes in N. crassa, it is imperative to know whether NCW-1 exerts its function on cellulase expression via CRE1. The results indicated that the Δncw-1Δcre-1 strain exhibited stronger protein production capability than either the Δcre-1 strain or the Δncw-1 strain (Fig. 2). The endoglucanase activity, β-glucosidase activity and secreted protein in the double deletion strain were 39, 44 and 77% higher than those of Δncw-1. Surprisingly, the xylanase activity of Δncw-1Δcre-1 increased to about 1.5-fold that of the reference strain Δncw-1. These results confirm that deleting both ncw-1 and cre-1 had a significant synergistic effect on lignocellulase production. On the other hand, a real-time PCR analysis indicated that the disruption of ncw-1 dramatically increased production of the cre1 transcript on Avicel medium (Supplementary Fig. 2). In conclusion, these data indicate that enhanced cellulase gene expression in the Δncw-1 is not the result of relief from catabolite repression mediated by cre-1.
Phenotype of Δcre-1Δncw-1 double mutant when grown on Avicel medium. Conidia from Δcre-1Δncw-1, Δcre-1, Δncw-1 and wild-type (WT) strains were separately inoculated into Avicel medium and batch cultured for 7 days. The total extracellular protein concentration (a), endoglucanase activity (b), β-glucosidase activity (c) and xylanase activity (d) were then measured. Values represent the means of at least three replicates, error bars show standard deviation
Deletion of NCW-1 improves cellobiose uptake in N. crassa
Based on previous findings, a N. crassa mutant carrying deletions of genes encoding three major β-glucosidases (Δ3βG) is an excellent system for dissecting the cellulase induction pathway (Znameroski et al. 2012). To test the effect of ncw-1 on cellulase gene expression under cellobiose condition, a Δ3βGΔncw-1 quadruple mutant was constructed by crossing. Cellobiose consumption and cellulase production assays were performed in Δ3βGΔncw-1 and Δ3βG strains. The consumption assay indicated that the Δ3βGΔncw-1 strain had dramatically increased cellobiose consumption compared with its parental strain. Cellobiose was almost depleted after 48 h of Δ3βGΔncw-1 cultivation, while there was still 6 g cellobiose/l in the culture of Δ3βG (Fig. 3a). Surprisingly, the Δ3βGΔncw-1 strain displayed a faster cellulase induction and a significant enhancement of cellulase production when transferred to cellobiose. The protein production of Δ3βGΔncw-1 was up to 141 μg/ml after 24 h induction, which was significantly higher than that of Δ3βG strain (below 10 μg/ml) (Fig. 3b).
Cellobiose consumption (a) and protein production (b) by the Δ3βGΔncw-1 strain on 1% (w/v) cellobiose medium. All strains were grown for 20 h on 2% (w/v) sucrose, followed by a transfer to 1% (w/v) cellobiose for 48 h. Values represent the means of at least three replicates, error bars show standard deviation
To observe the role of ncw-1 in cellulase expression, transcriptomic analysis of Δ3βGΔncw-1 strain on cellobiose was performed using Δ3βG as the control. Consistent with the cellulase production phenotype on 1% (w/v) cellobiose, the cellulase genes (NCU07340, NCU09680 and NCU00762), hemicellulase genes (NCU02855, NCU05955 and NCU07326), lytic polysaccharide monooxygenase genes (NCU08760, NCU02916, NCU02240 and NCU01050) and the cellobiose dehydrogenase gene (NCU00206) have significantly higher expression levels in Δ3βGΔncw-1 (Fig. 4a). Compared with Δ3βG, the expression level of cdt1 and cdt2 in Δ3βGΔncw-1 were up-regulated 19- and 15-fold, respectively (Fig. 4b), further showing that cellobiose uptake is dramatically accelerated in Δ3βGΔncw-1 consistent with more efficient transport of cellobiose into the cell. However, a detailed mechanistic description of how the ncw-1 influences cdt gene expression requires further study.
Transcriptome analysis of the Δ3βGΔncw-1 strain on cellobiose. a The transcriptional pattern of CAZy genes responding to 1% (w/v) cellobiose in Δ3βGΔncw-1 and Δ3βG strains. The size of dots represents the fold change between Δ3βGΔncw-1 and Δ3βG. Lytic polysaccharide monooxygenases (AA, red points), Carbohydrate esterases (CE, blue points), Glycosyltransferases (GT, purple points), Glycoside hydrolases (GH, green points), Polysaccharide lyases (PL, orange points). Black points represent genes with no differences in the expression. cbh-1 (NCU07340), gh6-2 (NCU09680), gh61-3 (NCU02916), gh5-1 (NCU00762), gh61-1 (NCU02240) and cdh-1 (NCU00206). b Expression patterns of the key transcriptional factors and cellodextrin transporters involved in cellulase induction in Δ3βGΔncw-1 and Δ3βG strains. TF transcription factor, TP transporter protein. All strains were grown for 20 h on 2% (w/v) sucrose, followed by a transfer to 1% (w/v) cellobiose for 4 h. Values represent the means of triplicates; error bars show standard deviation
Cellulase- and hemicellulase-related transcriptional factors clr1, clr2 and xlr1 were also up-regulated twofold, 17-fold and sevenfold in Δ3βGΔncw-1 on 1% (w/v) cellobiose, respectively (Fig. 4b). Likewise, the transcription factor vib-1, which is essential for cellulase production and is involved in repressing both glucose signaling and CCR (Xiong et al. 2014), was also up-regulated sixfold. Meanwhile, several cellulase repressor genes were significantly down-regulated. The N. crassa rca-1 (NCU01312) was down-regulated 14-fold, deletion of this gene significantly improves lignocellulase production when grown on plant biomass (Wang et al. 2015). In addition, the transcription factor res-1, which is required for endoplasmic reticulum stress response during cellulase synthesis (Fan et al. 2015), was down-regulated 3.8-fold. The most significant change in expression occurred with ada-6, which was down-regulated 51-fold in the hypercellulase-producing strain Δ3βGΔncw-1. This result accords with our unpublished data that disruption of ada-6 increases cellulase production by threefold in N. crassa. Taken together, the cellulase expression machinery was dramatically stimulated in Δ3βGΔncw-1. Overall, our findings suggest that NCW-1 affects cellulase induction principally by increasing uptake of cellobiose, a principal inducer of cellulase production in N. crassa and other cellulolytic filamentous fungi.
In conclusion, N. crassa ncw-1 is involved in the cellulase induction pathway. The improved cellulase production produced by deleting ncw-1 is not a consequence of relief from CCR of cre1 pathway, which suggests a novel strategy for improving cellulase production by co-disruption of ncw-1 and cre-1. Furthermore, NCW-1 could suppress cellulase induction via influencing cellulase inducer uptake. Thus, the characterization of ncw-1 in this study provides new insights into the cellulase induction pathway in filamentous fungi and may facilitate industrial strain improvement for cellulase production.
References
Fan F, Ma G, Li J, Liu Q, Benz JP, Tian C, Ma Y (2015) Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol Biofuels 8:66
Li J, Lin L, Li H, Tian C, Ma Y (2014) Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides d-glucose, d-xylose and l-arabinose. Biotechnol Biofuels 7:31
Lin L, Fang W, Liao X, Wang F, Wei D, St Leger RJ (2011) The MrCYP52 cytochrome P450 monoxygenase gene of Metarhizium robertsii is important for utilizing insect epicuticular hydrocarbons. PLoS One 6:e28984
Phillips CM, Iavarone AT, Marletta MA (2011) Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J Proteom Res 10:4177–4185
Sun Y, Cheng J (2002) Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83:1–11
Sun J, Glass NL (2011) Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One 6:e25654
Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, Glass NL (2009) Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA 106:22157–22162
Wang B, Cai P, Sun W, Li J, Tian C, Ma Y (2015) A transcriptomic analysis of Neurospora crassa using five major crop residues and the novel role of the sporulation regulator rca-1 in lignocellulase production. Biotechnol Biofuels 8:21
Wosten HA, Moukha SM, Sietsma JH, Wessels JG (1991) Localization of growth and secretion of proteins in Aspergillus niger. J Gen Microbiol 137:2017–2023
Xiong Y, Sun J, Glass NL (2014) VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PLoS Genet 10:e1004500
Zhang Y, Zhang J, Xiao P, Wang T, Qu Y (2012) Improved cellulase production via disruption of PDE01641 in cellulolytic fungus Penicillium decumbens. Bioresour Technol 123:733–737
Zhang W, Kou Y, Xu J, Cao Y et al (2013) Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J Biol Chem 288:32861–32872
Zhou Q, Xu J, Kou Y, Lv X, Zhang X, Zhao G, Zhang W, Chen G, Liu W (2012) Differential involvement of beta-glucosidases from Hypocrea jecorina in rapid induction of cellulase genes by cellulose and cellobiose. Eukaryot Cell 11:1371–1381
Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, Cate JH, Glass NL (2012) Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc Natl Acad Sci USA 109:6012–6017
Znameroski EA, Li X, Tsai JC, Galazka JM, Glass NL, Cate JH (2014) Evidence for transceptor function of cellodextrin transporters in Neurospora crassa. J Biol Chem 289:2610–2619
Acknowledgements
We thank Dr. Raymond J St. Leger from University of Maryland, College Park for revising the manuscript. This work was supported by the National Natural Science Foundation of China (31501007, 31471186) and Tianjin Zhuanxiang Grant 11ZCZDSY08400.
Supporting information
Supplementary Table 1—Primers used in this study.
Supplementary Table 2—Relative expression level of all genes detected in Δ3βG and Δ3βGΔncw-1 on 1 % (w/v) cellobiose.
Supplementary Fig. 1—Hyphal morphologies of Δncw-1 and wild type strains.
Supplementary Fig. 2—The expression levels of cellulase genes and transcriptional factor cre-1 in Δncw-1 strain monitored by quantitative RT-PCR after grown on Avicel for 2 days.
Author information
Authors and Affiliations
Corresponding author
Additional information
Liangcai Lin and Yong Chen have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lin, L., Chen, Y., Li, J. et al. Disruption of non-anchored cell wall protein NCW-1 promotes cellulase production by increasing cellobiose uptake in Neurospora crassa . Biotechnol Lett 39, 545–551 (2017). https://doi.org/10.1007/s10529-016-2274-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2274-1