Abstract
Objective
To elucidate the molecular mechanism of microRNA-215 (miR-215) in the migration and invasion of high grade glioma.
Results
42 Patients were analysed for clinicopathological characteristics. qRT-PCR showed that miR-215 was up-regulated in glioma tissues compared with non-neoplastic brain tissues (P < 0.05). The up-regulated miR-215 was closely associated with high grade glioma (P < 0.01) and poor overall survival (P < 0.01). Transwell assay showed that re-expression of miR-215 enhanced migration and invasion of glioma cells. miR-215 also down-regulated retinoblastoma tumor suppressor gene 1 (RB1) expression by targeting its 3′-UTR. Reversely, re-expression of RB1 inhibited partial effect of miR-215 on migration and invasion in vitro.
Conclusions
Re-expression of miR-215 promoted cell migration and invasion of glioma by targeting RB1. miR-215 can thus be used as a biomarker for tumor progression and prognosis in human high grade glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gliomas are a heterogeneous group of neoplasias that account for the majority of primary tumors of the central nervous system (Duan et al. 2015). Higher WHO-grade glioma (III + IV) carries a dismal prognosis of less than a 2 year median survival (Stupp et al. 2005). A major determinant of the high lethality of high-grade glioma is its diffuse invasion into healthy brain tissue thus precluding complete surgical resection (Le et al. 2015). Besides, glioma cells are found actively migrating throughout the extracellular spaces of the brain, nearly in the same way as embryonic neurons and glia cells migrate along preferred extracellular routes in the developing brain (Cuddapah et al. 2014). Despite advances in treatments following surgical resection, the overall survival remains poor. Therefore, it is necessary to understand molecular mechanism of glioma and identify underlying biomarkers.
MicroRNAs (miRNAs) are endogenous, small non-coding RNA sequences that are abundant in extracellular exosomes. They can be transferred from cell to cell by exosome release and uptake, resulting in cross-cellular gene-regulation (Hu et al. 2012). More than 1000 different miRNAs have been identified in humans according to the miRBase (Kozomara and Griffiths-Jones 2011). Aberrant expression of miRNA is involved in tumor progression, such as proliferation, invasion, migration and angiogenesis. In glioma analysis, many miRNAs, including miR-215 (Tong et al. 2015), miR-21 (Gabriely et al. 2008) and miR-218 (Tu et al. 2013), are dysregulated. In particular, miR-215 has been studied in various human malignancies. In gastric cancer, it is up-regulated and is a potential biomarker for prognosis (Deng et al. 2014). miR-215 is strongly up-regulated in human glioma cells and is involved in TGF-β1-induced oncogenesis (Tong et al. 2015). However, the function of miR-215 in regulating glioma progression is poorly understood.
Retinoblastoma tumor suppressor gene 1 (RB1), located on 13q14.1-q14.2, is a key regulator to control the G1/S transition during cell cycle progression by interacting with the E2F transcription factor family proteins. It is a tumor suppressor protein that is dysfunctional in several major cancers, including osteosarcoma (Deshpande and Hinds 2006), cervical carcinoma (Doorbar 2006), liver cancer (Munakata et al. 2007) and gastric cancer (Cito et al. 2010). In this study, we have explored the relationship between miR-215 and RB1 in high-grade glioma. Our data show that miR-215 is strongly up-regulated in glioma tissues compared with control tissues. Re-expression of miR-215 significantly promoted cell migration and invasion of glioma cells and, furthermore, RB1 was a potential target of miR-215. Collectively, these results indicate that miR-215 enhances glioma cell migration and invasion by regulating RB1 in high grade glioma.
Materials and methods
Patients and tissue samples
This study was approved by the Ethics Committee of Qilu Hospital of Shandong University, Shandong, P. R. China. All specimens were handled and made anonymous according to the ethical and legal standards, and written informed consent was obtained from all patients prior to sample collection.
42 patients were involved in the study between January 2010 and December 2015, and the patients’ clinicopathological characteristics were analyzed. Glioma tissues and non-neoplastic brain tissues were collected. Each sample was morphologically evaluated according to the WHO. Tissues were immediately cut and snap-frozen in liquid N2 before being stored at −80 °C for RNA extraction.
Cell lines
Human glioma cell lines (U87MG and U251MG) and HET293T cells were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) FBS.
Transfection
The transfection of RB1 expression vector was performed using Lipofectamine LTX (Invitrogen), and RNA-related vectors were transfected to cells using Lipofectamine RNAiMAX.
Quantitative Real-time PCR
Total RNA was extracted from tissues using Trizol. RNA was subsequently treated with RNase-free DNase I (Roche). Reverse-transcribed complementary DNA was synthesized from RNA using High Capacity cDNA Synthesis Kit (Applied Biosystems, Carlsbad, CA, USA), and quantitative RT–PCR (qRT-PCR) was performed with SYBR Premix ExTaq (TaKaRa, Dalian, China) with the Roche LightCycler 480 system (Roche). The relative expression of miR-215 was calculated by 2−△△CT method, normalized against the endogenous snRNA U6 control. The qRT-PCR for RB1 expression was similar. The relative expression of RB1 was calculated by 2−△△CT method and normalized to reduced glyceraldehyde-phosphate dehydrogenase (GAPDH).
Western blotting
Cells were treated with high KCl lysis buffer (10 mM Tris/HCl, pH 8, 140 mM NaCl, 300 mM KCl, 1 mM EDTA, 0.5% Triton X-100, and 0.5% sodium deoxycholate) supplemented with protease inhibitor cocktail (Roche) for a complete lysis. Protein concentrations were determined by BCA Protein Assay Kit (Pierce, Holmdel, NJ, USA). Cell protein extracts were separated by 10% SDS-PAGE and electroblotted onto a nitrocellulose membrane. Membranes were blocked with 5% (w/v) BSA in Tris-Buffered Saline with Tween 20 (TBST) buffer and incubated with primary antibody, followed by incubation with appropriate secondary antibody at room temperature. Specific proteins were detected using the enhanced chemiluminescence system (GE Healthcare). Antibodies against RB1 and GAPDH were purchased from Sigma–Aldrich. GAPDH was used for internal reference.
Luciferase reporter assay
The RB1 3′-UTR was amplified by PCR from genomic DNA and inserted into psiCHECK-2 vector. The predicted miR-215 binding sites were mutated using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). MiR-215 mimic and psiCHECK-2 Report vectors containing WT or MUT 3′-UTR of RB1 were transfected into HEK293T cells. After 48 h, the cells were harvested and assayed using Dual Luciferase Assay (Promega). Assays were read in a Veritas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA, USA). Firefly luciferase signal was normalized to renilla luciferase signal.
In vitro migration and invasion assay
Cells (5 × 105) were placed on the top of polycarbonate Transwell filters (without Matrigel for migration assay) or plated on the top of Matrigel-coated polycarbonate Transwell filter (for invasion assay) in the top chamber of the QCM 24-Well Cell Invasion Assay (Cell Biolabs,San Diego, CA, USA). Medium supplemented with serum was used as a chemoattractant in the bottom chamber. The cells were suspended in the serum-free medium and incubated at 37 °C for 8 h (migration assay) or 48 h (invasion assay). The non-migratory or non-invasive cells in the top chambers were removed with cotton swabs. The migrated and invaded cells on the lower membrane surface were fixed in 100% methanol for 10 min, air-dried, then stained with 4′,6-diamidino-2-phenylindole (DAPI) and counted under a microscope.
Statistical analysis
All statistical analyses were performed using SPSS 16.0 software (SPSS, Chicago, IL, USA). Differences between variables were assessed using the χ2 test unless otherwise noted. Measurement data were analyzed using Student’s t test. The Kaplan–Meier method was applied to calculate data from the survival curves. All data presented in this study have been repeated at least three times from three independent experiments and are presented as mean ±SD. Differences were considered significant at P-values < 0.05.
Results
MiR-215 expression is increased in high grade glioma
We examined the expression of miR-215 in 42 pairs of glioma tissues and non-neoplastic brain tissues by qRT-PCR. Expression of miR-215 in glioma tissues was significantly increased compared with that in non-neoplastic brain tissues (P < 0.05) (Fig. 1a).
MiR-215 is up-regulated in high-grade glioma and is associated with disease progression. a miR-215 expression detection in 42 pairs of glioma tissues and non-neoplastic brain tissues. b Up-regulation of miR-215 shown to be associated with a poor 5-year overall survival rate. Patients were divided into two groups according to the median miR-215 expression in glioma. *P < 0.05
The relationship between miR-215 expression and clinicopathological characteristics in 42 patients was analyzed. Increased expression of miR-215 was significantly correlated with high-grade glioma (III + IV) (P < 0.01) (Table 1). No significance was found in age, gender or KPS score.
To understand the prognostic significance of miR-215 upregulation in high-grade glioma, we analyzed the correlation between miR-215 expression and patient’s 5-year survival. We found that up-regulation of miR-215 was associated with a poor 5-year overall survival (OS) rate (P < 0.01) (Fig. 1b).
Re-expression of miR-215 promotes migration and invasion of glioma cell in vitro
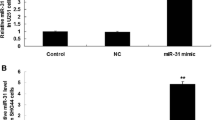
Given that the expression of miR-215 is associated with cell metastasis, we hypothesized that miR-215 play a key role in carcinogenesis and progression of glioma. To better understand the biological function of miR-215 in the development of glioma, we transfected miR-215 into U87MG cells (Fig. 2a). As detected by Transwell assays, the re-expression of miR-215 in U87MG cells remarkably enhanced cell migration and invasion compared with miR-control cells (Fig. 2b).
miR-215 down-regulates RB1 expression by directly targeting its 3′-UTR
To explore downstream targets of miR-215, bioinformatics analysis was performed using two online algorithms, TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do). RB1 was identified as a putative target gene, and one binding site was found from 1582 to 1589 bp, which was predicted highly conserved. A mutation of 3′-UTR of RB1 at binding site was constructed using the QuikChange Mutagenesis Kit at the binding site, named as MUT (Fig. 3a). To confirm the predictions, a luciferase reporter assay was performed in HEK293T cells. As shown in Fig. 3b, the relative luciferase activities were considerably decreased in cells co-transfected with miR-215 and 3′-UTR-WT luciferase reporter compared with that in cells co-transfected with miR-control and 3′-UTR-WT luciferase reporter. Inhibition was abolished in cells co-transfected with mutant RB1 3′-UTR (MUT) luciferase reporter and miR-215.
MiR-215 directly targets the 3′-UTR of RB1. a The putative binding site in the 3′-UTR of RB1. Mutation was generated in the complementary sites for the seed regions in miR-215. b Analysis of luciferase activity. HEK293T cells were co-transfected with the miR-215 mimic and a firefly luciferase reporter plasmid containing MT or MUT 3′-UTR of RB1. Firefly luciferase activity was normalized to Renilla luciferase activity. c Re-expression of miR-215 in U87MG cells significantly decreased the RB1 expression at both mRNA and protein levels. d The silence of miR-215 increased the expression of RB1 at both mRNA and protein levels. *P < 0.05
qRT-PCR and western blot assays were performed to detect the functional target of miR-215. The re-expression of miR-215 significantly reduced RB1 expression at mRNA and protein levels in U87MG cells (Fig. 3c). And the inhibition of miR-215 increased the expression of RB1 expression at mRNA and protein levels in U251MG cells (Fig. 3d).
Re-expression of RB1 attenuates the miR-215 induced promotion of migration and invasion in vitro
To confirm the effect of RB1 on migration and invasion induced by miR-215 in glioma, plasmids expressing RB1 or control vector were transfected into U87MG cells, which contsining miR-215 mimic. The expression of RB1 was detected by q-RT PCR and Western blot (Fig. 4a).
Compared with cells co-transfected with control and miR-215, the expression of RB1 was dramatically decreased in cells co-transfected with RB1 and miR-215 at mRNA and protein levels. We further examined the effect of RB1 on migration and invasion in cells co-transfected with miR-215 and RB1 expression plasmids or control vectors. As shown in Fig. 4b, re-expression of RB1 significantly attenuated the promotion of migration and invasion compared with U87MG (control vector + miR-215).
Discussion
Cancer development is a complex process which requires transcription and posttranscriptional regulation of gene expression. miRNAs play a key role in cancer progression by targeting genes (Zhang et al. 2012). miR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to Hypoxia by targeting KDM1B (Hu et al. 2016). In this study, we have shown that miR-215 was strongly up-regulated in glioma tissues compared with non–neoplastic brain tissues and that miR-215 regulates glioma cell migration and invasion. Besides, miR-215 bound to the 3′-UTR of RB1 and regulated its expression at mRNA and protein levels. In addition, RB1 is found involving in regulating the migration and invasion of glioma cells.
MiR-215 was first identified as a tumor suppressor by regulating a large number of genes that regulate cell cycle progression (Georges et al. 2008; Pichiorri et al. 2010). Generally, miR-215 is down-regulated in cancers, such as colon tumor (Karaayvaz et al. 2011), nephroblastoma (Senanayake et al. 2012), colorectal cancer (Li et al. 2013) and esophageal adenocarcinoma (Wijnhoven et al. 2010). In contrast, miR-215 is preferentially up-regulated in gastric cancer (Deng et al. 2014; Li et al. 2016) and glioma (Tong et al. 2015). Here, our data revealed that miR-215 was significantly increased in the glioma tissues compared with control samples. Increased expression of miR-215 was significantly associated with poor 5-year overall survival. And up-regulation of miR-215 dramatically enhanced cell migration and invasion in glioma cells. This promotion effect was achieved partially via targeting RB1. Western blotting showed that miR-215 mimic reduced RB1 expression and inhibition of miR-215 increased RB1 expression.
RB1 is involved in cell cycle suppression by preventing its progression from the G1 to S phase of the cell division cycle (Goodrich et al. 1991). In addition, RB1 could inhibit transcription factors of the E2F family, which are composed of dimers of an E2F protein and a dimerization partner protein (Wu et al. 1995). The pRB, hypophosphorylated state RB1, release the restriction of progression from the G1 phase to the S phase of the cell cycle. The initial phosphorylation is performed by cyclin D/CDK4/CDK6 and followed by additional phosphorylation by cyclin E/CDK2 (Munger and Howley 2002). RB1 pathways via p14ARF may regulate p53, which has many mechanisms of anticancer function and plays a role in apoptosis, genomic stability, and inhibition of angiogenesis (Hu et al. 2007). In this study, RB1 was involved in the pathology of glioma. It was directly regulated at the mRNA and protein levels by miR-215. Re-expression of RB1 attenuated the miR-215 induced promotion of migration and invasion in vitro, indicating that RB1 functions as a tumor suppressor in glioma.
There are some limitations in this study. Patients with glioma were divided into high-grade glioma group (grade III and IV) and low-grade glioma group (grade I and II) and differences were not studied between grade III and IV. In future studies, greater effort will be taken to explore the different molecular mechanism between grade III and IV of glioma.
In summary, miR-215 was dramatically increased in human glioma and up-regulation of miR-215 correlated with poor prognosis and high-grade glioma. Re-expression of miR-215 promoted cell migration and invasion of glioma by targeting RB1. Taken together, miR-215 can be used as a biomarker for tumor progression and prognosis in human glioma.
Change history
23 July 2022
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s10529-022-03283-6
References
Cito L, Pentimalli F, Forte I, Mattioli E, Giordano A (2010) Rb family proteins in gastric cancer (review). Oncol Rep 24:1411–1418
Cuddapah VA, Robel S, Watkins S, Sontheimer H (2014) A neurocentric perspective on glioma invasion. Nat Rev Neurosci 15:455–465
Deng Y, Huang Z, Xu Y, Jin J et al (2014) MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett 342:27–35
Deshpande A, Hinds PW (2006) The retinoblastoma protein in osteoblast differentiation and osteosarcoma. Curr Mol Med 6:809–817
Doorbar J (2006) Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 110:525–541
Duan R, Han L, Wang Q, Wei J, Chen L, Zhang J, Kang C, Wang L (2015) HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-beta pathways. Oncotarget 6:27778–27793
Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28:5369–5380
Georges SA, Biery MC, Kim SY, Schelter JM et al (2008) Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res 68:10105–10112
Goodrich DW, Wang NP, Qian YW, Lee EY, Lee WH (1991) The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell 67:293–302
Hu W, Feng Z, Teresky AK, Levine AJ (2007) p53 regulates maternal reproduction through LIF. Nature 450:721–724
Hu G, Drescher KM, Chen XM (2012) Exosomal miRNAs: biological properties and therapeutic potential. Front Genet 3:56
Hu J, Sun T, Wang H, Chen Z et al (2016) MiR-215 is induced post-transcriptionally via HIF-drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell 29:49–60
Karaayvaz M, Pal T, Song B, Zhang C, Georgakopoulos P, Mehmood S, Burke S, Shroyer K, Ju J (2011) Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer 10:340–347
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Le AP, Huang Y, Pingle SC, Kesari S, Wang H, Yong RL, Zou H, Friedel RH (2015) Plexin-B2 promotes invasive growth of malignant glioma. Oncotarget 6:7293–7304
Li S, Gao J, Gu J, Yuan J, Hua D, Shen L (2013) MicroRNA-215 inhibits relapse of colorectal cancer patients following radical surgery. Med Oncol 30:549
Li N, Zhang QY, Zou JL, Li ZW et al (2016) miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget 7:4817–4828
Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM (2007) Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog 3:1335–1347
Munger K, Howley PM (2002) Human papillomavirus immortalization and transformation functions. Virus Res 89:213–228
Pichiorri F, Suh SS, Rocci A, De Luca L et al (2010) Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 18:367–381
Senanayake U, Das S, Vesely P, Alzoughbi W, Frohlich LF, Chowdhury P, Leuschner I, Hoefler G, Guertl B (2012) miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis 33:1014–1021
Stupp R, Mason WP, van den Bent MJ, Weller M et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996
Tong YQ, Liu B, Zheng HY, Gu J et al (2015) MiR-215, an activator of the CTNNBIP1/beta-catenin pathway, is a marker of poor prognosis in human glioma. Oncotarget 6:25024–25033
Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin W, Zhang Y (2013) MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res 73:6046–6055
Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ, South Australian Oesophageal Research G (2010) MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. Br J Surg 97:853–861
Wu CL, Zukerberg LR, Ngwu C, Harlow E, Lees JA (1995) In vivo association of E2F and DP family proteins. Mol Cell Biol 15:2536–2546
Zhang Y, Dutta A, Abounader R (2012) The role of microRNAs in glioma initiation and progression. Front Biosci 17:700–712
Author information
Authors and Affiliations
Corresponding author
Additional information
Yuzhen Wei and Jianjing Sun have equally contributed to this work.
About this article
Cite this article
Wei, Y., Sun, J. & Li, X. RETRACTED ARTICLE: MicroRNA-215 enhances invasion and migration by targeting retinoblastoma tumor suppressor gene 1 in high-grade glioma. Biotechnol Lett 39, 197–205 (2017). https://doi.org/10.1007/s10529-016-2251-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2251-8