Abstract
Stable carriers are required for delivering siRNA to cells. The use of polyethyleneimine (PEI) as gene carrier has been researched extensively; however, it does not provide sufficient protection from RNase degradation and is not suitable for targeted siRNA delivery to specific cells. In this study, two repeats of Fc binding domain of protein G (C2) were used to introduce a specific antibody to PEI-based carrier of siRNA. In addition, we used the double-stranded RNA binding domain (DRBD) that can bind to siRNA. The complex, consisting of PEI, siRNA and constructed fusion protein, TrxC2DRBD including C2 and DRBD domains, could protect siRNA from RNase degradation. Furthermore, cell specific siRNA delivery into HeLa cells could be performed by the complex fusion with specific antibodies via C2 domain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Small interfering RNA is a short double-stranded RNA, 20–25 nucleotides in length that induces gene silencing called RNA interference (RNAi). RNAi involves small RNAs, such as short-interfering RNA (siRNA) and microRNA (miRNA). siRNA is the most widely researched gene silencing therapeutic application (Wang et al. 2010). However, siRNA has a strong negative charge so it is difficult to deliver into cells. To overcome this problem, polyethyleneimine (PEI) has been researched in depth because of its strong cationic charge, which is due to multiple amines. PEI is used as a gene carrier for gene transfections (Boussif et al. 1995). Large branched PEIs, such as PEI 25K (average MW 25,000), have been well studied as gene carriers (Hattori and Maitani 2007). This property gives it a proton sponge effect that is expected to greatly increase the transfection efficiency (Tan et al. 2011).
siRNA has a low silencing effect and so high siRNA concentrations are required. However, high concentrations of siRNA or PEI cause toxicity. To overcome this problem, Lee et al. (2010) developed poly-siRNA/PEI complexes. Their complexes showed a high gene silencing effect. Kong et al. (2011) developed a multimerized siRNA carrier with nanoparticles and a 25 kDa PEI. Their multimerized siRNA carrier was stable and compact and showed a high gene silencing efficiency.
Another problem is the degradation that siRNA can undergo by RNase. Mao et al. (2006) reported that PEI alone, which can form siRNA/PEI complexes, did not provide sufficient protection from RNase degradation. They therefore developed a PEI-graft-PEG block copolymer, which protected siRNA to a greater extent than siRNA/PEI.
Additionally, PEI is not suitable for gene delivery because it does not have any targeting specificity. In order to achieve specific delivery, antibodies have been conjugated to PEI. In one application, Strehblow et al. (2005) developed an efficient and cell-specific non-viral gene delivery system using monoclonal antibodies coupled with branched 25 kDa PEI. Han et al. (2008) also reported efficient and specific delivery of PEI conjugated with antibodies (PEI-anti-MMP-2) against matrix metalloproteinase 2 (MMP-2), a surface marker protein on cancer cells. Conjugation with an antibody could provide target specificity for PEI gene delivery system. However, this method requires chemical modification of the antibody, which is a complicated process that may affect the binding affinity or specificity of the antibody. To overcome these problems, we have made the system applicable to gene delivery using any antibody and a fusion protein containing two repeats of the C domain (C2).
In this study, we have focused on the double-stranded RNA binding domain (DRBD) from the RNA-activated protein kinase that binds to double-stranded RNA. DRBD requires a minimum of 16 base-pairs of dsRNA for binding (Bevilacqua and Cech 1996). This means that DRBD can bind to and thus protect siRNA. The fusion protein TrxC2DRBD was engineered with the expectation that it would bind to siRNA and protect it from RNase degradation. This was then confirmed.
Materials and methods
Materials
The plasmid pUC 18 cloning vector was obtained from Toyobo (Japan). Plasmid pET-32c and Escherichia coli BL21 (DE3) were purchased from Novagen. Synthesized DNA fragments were purchased from Hokkaido system science (Japan). The restriction enzymes and ligase were purchased from Takara Bio (Japan). Human cervical HeLa cell was obtained from the Riken Cell Bank (Tsukuba, Japan). All other chemicals were of analytical grade.
Construction of plasmids
The plasmid pET-TrxC2 was constructed as follows. The gene encoding the immunoglobulin G (IgG) Fc binding domain (C2) of protein G was digested with BamHI and HindIII from the pBS-G2 vector (Tanaka et al. 2006) and the resulting fragment was inserted into the pET-32c vector for addition of Trx-tag and His-tag at 5′-end.
The plasmid pET-His-DRBD was constructed as follows. DRBD gene, a part of human PKR DRBD-1, was amplified from cDNA of HeLa cell by PCR using primers (5′-CCATGGCTCGAGAGATCTCATCAGCAGGTTTCTTCATGGA and GCGGCCGCTCAAGGACTAACTGCCTTCTTT). The amplified fragment was inserted into a pUC18 vector digested with SmaI. The resulting plasmid, pUC18-DRBD, was digested with NcoI and NotI, and the fragment was inserted into pET-His-C2 digested with same restriction enzymes.
The pET-His-C2-DRBD was constructed as follows. pUC 18-DRBD was digested with BglII and NotI and then inserted into pET-His-C2 digested with same restriction enzymes.
To add the Trx tag, pET-His-DRBD and pET-His-C2-DRBD were digested with NcoI and NotI and then inserted into pET32c digested with same restriction enzymes. These constructed plasmids were pET-Trx-DRBD, pET-Trx-C2-DRBD.
Expression and purification of fusion protein
To express the fusion proteins, E. coli BL21 (DE3) was transformed with the expression plasmids. Transformed cells were grown at 37 °C in LB medium with 100 μg ampicillin/ml to an OD660 value of 0.6–0.7. After inducing protein expression with IPTG at 1 mM, the cells were shaken for overnight at 25 °C. The cells were then harvested by centrifugation and re-suspended in Bugbuster (Novagen) reagent distilled with PBS (150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4, pH 7.4) with added 10 % Benzonase Nuclease (Novagen). After rotating for 30 min, the samples were centrifuged. Supernatants were applied to a His-Select Nickel Affinity Gel (Sigma) and rotated for 1 h at 4 °C. the gel was then washed with Ni–NTA buffer (20 mM phosphate, 0.5 mM NaCl) containing imidazole. After washing, the bound protein was eluted with Ni–NTA buffer (20 mM phosphate, 0.5 mM NaCl) containing 200 mM imidazole. The eluate was dialyzed twice for 90 min each against PBS. The purified proteins were then run on SDS-PAGE to confirm their molecular weights and their concentrations were measured using a BCA assay kit (Pierce).
Cell culture
Human cervical HeLa cells were grown in DMEM medium with 10 % (v/v) fetal bovine serum (FBS) and antibiotics (100 U penicillin/ml, 100 μg streptomycin/ml).
Construction of a cell line with stable GFP expression
Human cervical HeLa cells were seeded in 35 mm dish at 80 % of plate and cultured for 24 h. pcDNA3.1-GFP (6,160 bp) was transfected with effectene transfection reagent (Qiagen). After 48 h incubation after transfection, the cells were treated with G418 sulfate for selection of stable transformed cells.
Western blotting for confirming C2 domain activity
The transformed E. coli lysates were separated by SDS-PAGE in reducing conditions followed by protein transfer to a PVDF membrane for Western blotting. The membrane was then blocked with 0.25 % Block Ace (DS Pharma Biomedical, Japan) in PBS buffer for 30 min. After washing with PBS-T buffer (PBS with 0.05 % Tween 20), alkaline phosphatase (ALP)-labeled secondary goat anti-rat antibody (EY laboratories) was added and the membrane was incubated for 1 h and then washed with PBS-T and added ALP substrates (Sigma Fast, TR/Naphthol AS-MX).
Optimization of siRNA concentration for silencing of GFP
HeLa-GFP cells were seeded in 96 well plates at 4,000 cells/well and cultured for 24 h in 100 μl DMEM medium containing 10 % (v/v) FBS and 1 % penicillin/streptomycin. Lipofectamine 2000 RNAi, 1 μl, was incubated with 50 μl opti-MEM (Invitrogen) for 5 min at room temperature and then incubated with siRNA-GFP (Silencer GFP siRNA, Applied Biosystems Ambion) at either 50, 100 or 500 nM for 30 min at room temperature. After incubation for 72 h the cells were washed with PBS. HeLa-GFP cells were observed by fluorescent microscopy.
Gel shift assay for confirming a formation of siRNA–PEI complex
A sample with 0.5 nM siRNA–GFP was incubated with PEI (25 kDa, Sigma) at 0, 0.005, 0.05, 0.5, 0.7, 1.0, 2.5, 5.0 and 7.0 mM for 30 min at room temperature. The siRNA-GFP complexes were then mixed with 3× loading buffer and separated by agarose gel (2 %) electrophoresis at 100 V for 40 min in TAE buffer.
Evaluating transfection efficiency of siRNA/PEI complexes
HeLa-GFP cells were seeded in 96 well plates at 4,000 cells/well and cultured for 24 h in 100 μl DMEM medium containing 10 % (v/v) FBS and 1 % penicillin/streptomycin. 0.5 μM siRNA-GFP (50 nM) was incubated with PEI at 2.5, 3.5, 4, 4.5 and 5 mM for 30 min at room temperature. Each sample was tranfected for 6 h then the medium was changed at 72 h from transfection. Cells were washed with PBS and cell viability was evaluated using a cell counting kit-8.
Gel shift assay for confirming DRBD binding ability to siRNA
The complexes of siRNA/DRBD and siRNA/C2DRBD were formed with 50 nM siRNA and proteins at molar ratio of 1:1, 1:2 and 1:10 each. The mixtures of siRNA and each protein was incubated at room temperature for 30 min, then samples were separated by electrophoresis.
RNase protection assay
siRNA (10 μM) were mixed with DRBD and C2DRBD protein at molar ratio of 1:1, 1:2 and 1:10 for 30 min at room temperature. Then, RNase A (0.007 units/μl) were added to siRNA/DRBD and siRNA/C2DRBD complexes and incubated at 37 °C for 30 min followed by agarose gel electrophoresis.
Gel shift assay for confirming siRNA/protein/PEI complexes
To form siRNA and protein complexes, siRNA (50 nM) and proteins (500 nM) were incubated for 30 min at room temperature. PEI at 3.5, 4 and 4.5 mM was then added and the mixture was held at room temperature for 10 min. Electrophoresis was performed as same as before.
Evaluation of silencing efficiency of siRNA/C2DRBD/PEI complex
HeLa-GFP cells were seeded in 96 well plates at 3,500 cells/well and cultured for 24 h in 100 μl DMEM medium containing 10 % FBS and 1 % penicillin/streptomycin. siRNA-GFP, 50 nM, was incubated with TrxC2DRBD protein at molar ratio of 1:10 for 30 min at room temperature. Then complex solution was mixed with 0.2 μg antibody (monoclonal anti-human HLA class I antigen antibody, Sigma) and incubated for 10 min at room temperature. As a negative control, rabbit anti-sheep IgG (Zymed) was also prepared in the same way. Finally, PEI was added and incubated for 10 min at room temperature. These complexes were added to cells and incubated for 6 h. Then medium was changed. After 72 h from transfection, the cells were washed with PBS and observed by fluorescent microscopy.
Results and discussion
Expression and purification of fusion proteins
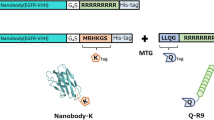
The strategy of this experiment is illustrated in Fig. 1a. Fragment C of protein G binds specifically to the constant region of immunoglobulin G (IgG). Therefore, antibodies may be changed easily by using this fusion protein. However, for using siRNA addresses the problem of its degradation by RNase, through construction of the TrxC2DRBD fusion protein. The DRBD domain of this protein binds to double stranded RNA and thus is expexted to protect siRNA from RNase.
a Strategy for specific siRNA delivery with the complex of antibody/fusion protein/PEI/siRNA. The fragment C2 from protein G binds specifically to the heavy chain constant region of immunoglobulin G (IgG). DRBD domain binds to double strand RNA for protection of siRNA from RNase. b Construction of fusion proteins
These fusion proteins were expressed in E. coli BL21 (DE3). The construction of fusion proteins is shown in Fig. 1b.
Purification of the fusion proteins was confirmed by SDS-PAGE (data not shown). Fusion proteins, TrxC2, TrxDRBD and TrxC2DR20, appeared at the expected molecular masses (34.8, 26.6 and 40.6 kDa). Both TrxC2 and TrxC2DRBD proteins were confirmed to retain IgG binding abilities by Western blotting (data not shown).
Optimization of siRNA concentration for silencing of GFP
The siRNA concentration was optimized using lipofectamine. In this experiment, siRNA were mixed with lipofectamine 2000 RNAi for 30 min at room temperature and then transfected to HeLa-GFP cells. Here, siRNA concentrations from 50 to 500 nM were tested. As shown in Fig. 2, all concentrations of siRNA showed the GFP silencing. Even 50 nM of siRNA silenced GFP. From this result, we concluded that the optimal siRNA concentration was 50 nM.
Evaluating transfection efficiency and cell viability assays of siRNA/PEI complexes
The concentrations of siRNA and PEI are important for gene silencing. High concentrations of siRNA and PEI are toxic to cells (Lee et al. 2010). To reduce the toxicity to cells, Jiang et al. (2009) developed a siRNA/PEI–HA complex.
Figure 3a shows the cell viability by adding siRNA/PEI complexes. The siRNA/PEI complex at 2.5 mM of PEI showed the highest cell viability. However, the siRNA/PEI complex at concentrations higher than 4 mM PEI showed low cell viability.
The siRNA transfection efficiency results (Fig. 3b) indicated that siRNA/PEI at 2.5 mM PEI complex showed the lowest transfection efficiency, which was almost the same as that for untreated cells. In contrast, siRNA/PEI at 4.5 and 5 mM PEI complex showed a high transfection efficiency, with the GFP silenced.
This result indicated that at high concentration PEI showed a high transfection efficiency but high concentrations of PEI also caused toxicity, especially at 4.5 and 5 mM. Low concentrations of PEI showed a low transfection efficiency compared to higher PEI concentrations, but had higher cell viability. Based on these results, a PEI concentration of 3.5 mM was used for further experiments.
RNase protection assays for siRNA, siRNA/DRBD and siRNA/C2DRBD complexes
One of the major problems associated with siRNA delivery is rapid enzymatic degradation. To overcome this problem, DRBD was fused to TrxC2. DRBD is expected to protect siRNA through its binding interactions. To confirm protection of siRNA by binding to DRBD, siRNA/protein complexes were treated with RNase (Fig. 4). Figure 4 indicates that upon addition of RNase A, siRNA without protein disappeared. On the other hand, siRNA with TrxDRBD and TrxC2DRBD proteins was stable as indicated by unchanged SDS-PAGE bands. These results showed that the DRBD protein protected siRNA from RNase A by binding to siRNA.
RNase protection assay of siRNA/TrxDRBD and siRNA/TrxC2DRBD complexes. 1 siRNA alone (RNase +), 2 siRNA:TrxDRBD molar ratio at 1:1(RNase +), 3 siRNA:TrxDRBD molar ratio at 1:2 (RNase +), 4 siRNA:TrxDRBD molar ratio at 1:10 (RNase +), 5 siRNA:TrxC2DRBD molar ratio at 1:1 (RNase +), 6 siRNA:TrxC2DRBD molar ratio at 1:2 (RNase +), 7 siRNA:TrxC2DRBD molar ratio at 1:10 (RNase +), 8 siRNA alone (RNase −)
Evaluation of the silencing efficiency of siRNA/C2DRBD/PEI complexes
After confirming triplex formation, complexes with antibodies were added to cells. Figure 5a shows the results of siRNA/TrxC2DRBD/PEI complex formation with or without added antibody. Here, the PEI was at 3.5 mM. The top column indicates transfection of siRNA/TrxC2DRBD/PEI in which some cells developed a murky GFP intensity. However, in the bottom column, results for transfection of siRNA/TrxC2DRBD/PEI/non-specific antibody complexes showed that there were some cells with almost the same GFP intensity. In contrast, siRNA/TrxC2DRBD/PEI/specific HLA antibody complexes, which are in the middle column of Fig. 5, clearly showed their silencing efficiency. Their GFP intensity became murky compared with siRNA/TrxC2DRBD/PEI complexes.
a Transfection efficiency of siRNA/TrxC2DRBD/PEI (3.5 mM), siRNA/TrxC2DRBD/PEI (3.5 mM)/specific HLA antibody and siRNA/TrxC2DRBD/PEI (3.5 mM)/non-specific antibody. b Cell viability of siRNA/lipofectamine, siRNA/PEI, siRNA/PEI/TrxC2DRBD, siRNA/PEI/TrxC2DRBD/specific HLA antibody and siRNA/PEI/TrxC2DRBD/non-specific antibody
Figure 5b shows the cell viability of siRNA/TrxC2DRBD/PEI with or without antibody complexes. The siRNA/TrxC2DRBD/PEI/specific antibody complex showed a high cell viability of 80 % compared with single cells and the same viability with siRNA/PEI complexes.
To conclude, siRNA/TrxC2DRBD/PEI complexes have as siRNA silencing effect and when they form specific antibody complexes these can have a greater silencing efficiency compared with siRNA/TrxC2DRBD/PEI. In addition, siRNA/TrxC2DRBD/PEI/specific antibody complexes had a high cell viability activity. These results indicated that engineered TrxC2DRBD proteins were useful as they had specific silencing efficiency and also protected siRNA.
Conclusion
TrxC2, TrxDRBD and TrxC2DRBD fusion proteins were designed to have a high gene silencing efficiency combined with specific delivery. We have focused on a siRNA delivery system using the C2 domain of protein G and DRBD. As siRNA can be degraded by enzymes, siRNA protection is important to achieve efficient siRNA delivery. The siRNA binding activity with DRBD was tested, and then the stability of this complex was evaluated using a protection assay. The fusion protein bound to and protected siRNA. Thus the siRNA/TrxC2DRBD/PEI/specific antibody complexes are useful due to their specific silencing efficiency and also because they protect siRNA.
References
Bevilacqua PC, Cech TR (1996) Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35:9983–9994
Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo : polyethtlenimine. Proc Natl Acad Sci USA 92:7297–7301
Han JY, Choi DS, Kim CH, Joo H, Min CK (2008) Selective gene transfer to endometrial cancer cells by a polymer against matrix metalloproteinase 2 (MMP-2). Cancer Biother Radiopharm 22:247–258
Hattori Y, Maitani Y (2007) Low-molecular-weight polyethyleneimine enhanced gene transfer by cationic cholesterol-based nanoparticle vector. Biol Pharm Bull 30:1773–1778
Jiang G, Park K, Kim J, Kim KS, Hahn SK (2009) Target specific intracellular delivery of siRNA/PEI-HA complex by receptor mediated endocytosis. Mol Pharm 6:727–737
Kong WH, Bae KH, Hong CA, Lee Y, Hahn SK, Park TG (2011) Multimerized siRNA cross-linked by gold nanoparticles. Bioconjugate Chem 22:1962–1969
Lee SY, Huh MS, Lee S, Lee SJ, Chung H, Park JH, Oh YK, Choi K, Kin¥m K, Kwon IC (2010) Stability and cellular uptake of polymerized siRNA (poly-siRNA)/polyethyleneimine (PEI) complexes for efficient gene silencing. J Control Release 141:339–346
Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Barkowsky U, Kissel T (2006) Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/siRNA polyplexes. Bioconjugate Chem 17:1209–1218
Strehblow C, Schuster M, Moritz T, Kirch HC, Opalka B, Petri JB (2005) Monoclonal antibody–polyethyleneimine conjugates targeting Her-2/neu or CD90 allow cell type-specific nonviral gene delivery. J Control Release 102:737–747
Tan SJ, Kiatwuthinon P, Roh YH, Kahn JS, Luo D (2011) Engineering nanocarriers for siRNA delivery. Small 7:841–856
Tanaka G, Funabashi H, Mie M, Kobatake E (2006) Fabrication of an antibody microwell array with self-adhering antibody binding protein G. Anal Biochem 350:298–303
Wang Y, Li Z, Han Y, Liang LH, Ji A (2010) Nanoparticle-based delivery system for application of siRNA in vivo. Curr Drug Metab 11:182–196
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bae, J., Mie, M. & Kobatake, E. Development of a specific siRNA delivery system into HeLa cells using an IgG-binding fusion protein. Biotechnol Lett 35, 2081–2089 (2013). https://doi.org/10.1007/s10529-013-1299-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-013-1299-y