Abstract
The 5′-thiolated DNA probe based on specific virulence gene, Omp85, was immobilized onto a screen-printed gold electrode followed by hybridization with 6–100 ng/6 μl (5.9 × 105–9.3 × 106 c.f.u.) of Neisseria meningitidis single stranded genomic DNA (ssG-DNA) for 10 min at 25 °C from the cerebrospinal fluid (CSF) of a meningitis patient. The Omp85 genosensor can detect as little as 6 ng ssG-DNA in 6 μl CSF of a human brain meningitis patient in 30 min including a response time of 1 min by cyclic voltammetry, differential pulse voltammetry (DPV) and electrochemical impedance. The sensitivity of the genosensor electrode was 2.6(μA/cm2)/ng using DPV with regression coefficient (R2) 0.954. The genosensor was characterized using Fourier transform infrared spectroscopy and atomic force microscopy. Omp85 genosensor was stable for 12 months at 4 °C with 12 % loss in DPV current.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meningitis is a severe inflammation in the meninges (outer membrane covering) of brain and spinal cord of human. Neisseria meningitidis is the main responsible pathogen causing human brain bacterial meningitis (Espinosa et al. 2001; Dash et al. 2012) that can lead to damage of the brain or even death of the patient. Meningitis is generally detected from patient CSF through microscopy (Dunbar et al. 1998), biochemical assays (Negi et al. 2010), latex agglutination test (Surinder et al. 2007), PCR (Kumar et al. 2011; Dash et al. 2012a), real-time PCR (RT PCR) (Staquet et al. 2007), microarray (Mahajan et al. 2009; Sethi et al. 2010; Patnaik et al. 2012) and biosensors (Patel et al. 2009, 2010). The present available methods are though cumbersome and have one or more limitations. Therefore, genosensors are gaining preference for diagnosis of the meningitis. Thiolated DNA has always been a better choice for genosensors because of their easy fabrication, economic value and ability to function at wide range of temperature, pH and solvents (Lin et al. 2011). Methylene Blue (MB) is widely used as redox indicator for electrochemical genosensors (Kumar et al. 2009, 2012).
Omp85 is a virulence gene that codes for a conserved outer membrane protein of N. meningitidis. Omp85 can be targeted for the development of a vaccine (Fitzpatrick and McInerney 2005). Omp85 also assists in effective insertion of lipids and integral proteins into the outer membrane of N. meningitidis (Voulhoux et al. 2003). A single stranded DNA probe was designed targeting Omp85 gene of N. meningitidis for specific hybridization on a screen-printed gold electrode.
Materials and methods
Chemicals and patient sample
The 5′-thiolated ssDNA probe (5′-CGTTTCCCAAGACAACCTGT-3′), based on our NCBI GenBank accession number HQ712171 (Dash et al. 2010), was synthesized by Innovision Medichem. SPGE (gold: counter and working electrode, silver: reference electrode) was procured from DropSens and modified at our Institute. All other chemicals used were of analytical grade. CSF from patients with suspected meningitis were obtained from National Centre for Disease Control (NCDC), Delhi.
Isolation of DNA from patient CSF
Patient CSF (0.5 ml) containing N. meningitidis was centrifuged at 16,000×g for 1 min. The upper viscous CSF was discarded and the pellet was suspended in 0.5 ml of TE (10 mM Tris/HCl, 1 mM EDTA) buffer, pH 8. The bacterial suspension was heated (for cell lysis) at 95 °C for 5 min and centrifuged at 16,000×g for 1 min. The amount of the genomic DNA (G-DNA) was quantified from the supernatant. In a second set of experiment, the N. meningitidis suspension was diluted serially and each dilution was plated in triplicate on Mueller–Hinton agar and grown at 37 °C for 18 h in CO2 rich environment. The experiment was repeated three times and the average c.f.u. of N. meningitidis corresponding to G-DNA was calculated for different dilutions. The supernatant containing G-DNA was denatured for 5 min at 95 °C to make single stranded DNA (ssDNA) of 6 ng/6 μl (5.9 × 105 c.f.u.), 12 ng/6 μl (1.2 × 106 c.f.u.), 25 ng/6 μl (2.4 × 106 c.f.u.), 50 ng/6 μl (4.6 × 106 c.f.u.) and 100 ng/6 μl (9.3 × 106 c.f.u.) for hybridization with the immobilized probe on modified SPGE. N.meningitidis, after infection in human CSF, doubles every 30 min.
Fabrication of genosensor
The working gold surface (12.6 mm2) was activated by treating with H2SO4/H2O2 (1:1, v/v) for 5 min and then washed with autoclaved Milli Q water followed by absolute ethanol subsequently with water and finally dried at room temperature 25 °C. The ssDNA-SH/Au was constructed by immobilizing 5 μl of 5′-thiolated Omp85 DNA probe (1 μM) onto SPGE for 24 h at 25 °C and the unbound DNA probe was removed by washing with PBS (50 mM phosphate buffer, 0.9 % NaCl), pH 7 and dried at room temperature for 3–4 h. The ssG-DNA 6–100 ng/6 μl from a patient CSF was hybridized at 25 °C for 10 min (optimized) with ssDNA-SH/Au. After hybridization, the electrode was washed 4–5 times with TE buffer, pH 8 to remove unhybridized N. meningitidis ssG-DNA and again washed with PBS, pH 7 before electrochemical study. The immobilization of ssDNA probe and hybridization with ssG-DNA was detected by CV, DPV and EI using FRA2 μAUTOLAB type iii, Metrohm. The overall experiment was repeated three times and the average values were taken for plotting graph between concentration of ssG-DNA and the oxidation peak current (Ip). The schematic fabrication of the genosensor is shown in Scheme 1.
The surface topography of both ssDNA-SH/Au and dsDNA-SH/Au electrodes (after hybridization) were characterized using FTIR in a frequency range of 400–4,000/cm and AFM in non-contact mode in a surface area range of 0.25 μm2 (5500SPM, Agilent Tech).
Results and discussion
CV studies
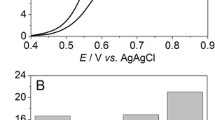
CV of ssDNA-SH/Au and dsDNA-SH/Au is shown in Fig. 1. The CV current of ssDNA-SH/Au (5.89 μA, curve a) was lower than that of bare gold (not shown in Fig. 1), due to higher rate of electron exchange between gold and Methylene Blue (MB). Whereas, strong association of MB with the unpaired nitrogenous bases (guanine) of ssDNA-SH/Au decreases the rate of electron exchange and hence decreases the current output. The oxidation current of dsDNA-SH/Au was 9.11 μA (curve b), 10.16 μA (curve c), 12.37 μA (curve d), 14.00 μA (curve e) and 14.69 μA (curve f) after hybridization with 6 ng/6 μl, 12 ng/6 μl, 25 ng/6 μl, 50 ng/6 μl and 100 ng/6 μl of ssG-DNA of N. meningitidis, respectively (Fig. 1). The oxidation peak current increased along with an increase in ssG-DNA was due to the presence of extra unhybridized guanine bases of G-DNA that interacted with MB molecules and resulted in increased current output. The hyperbolic curve between relative peak current (with respect to probe) Ip and concentration of ssG-DNA (Fig. 1 inset) shows that, the oxidation current increased up to 50 ng/6 μl of ssG-DNA. On further increasing the concentration of hybridizing DNA, the peak current does not increased further because all the immobilized ssDNA probe of gold surface (12.6 mm2) get saturated at 50 ng/6 μl and does not available for further hybridization with ssG-DNA. The genosensor can detect as low as 6 ng/6 μl ssG-DNA in CSF and the sensitivity of the electrode was 0.909 (μA/cm2)/ng with R2 0.94.
Cyclic voltammetry of (a) ssDNA-SH/Au and (b–f) hybridization with 6, 12, 25, 50 and 100 ng/6 μl of N. meningitidis ssG-DNA at 50 mV/s using 1 mM MB in 50 mM PBS, pH 7. The inset shows linear increase of the relative peak current Ip up to 50 ng/6 μl of ssG-DNA of N. meningitidis used for hybridization following equation I (μA) = 0.109 (μA/ng) × [ssDNA] (ng) + 2.983 (μA). 1 c.f.u. of N. meningitidis corresponds to 1.0 × 10−5 ng of ssG-DNA
DPV studies
The DPV current of ssDNA-SH/Au (35.78 μA, curve a) was lower than that of bare gold (not shown in Fig. 2). The current of dsDNA-SH/Au increased along with an increase in concentration of ssG-DNA of N. meningitides: 42.28 μA (6 ng/6 μl, curve b), 47.32 μA (12 ng/6 μl, curve c), 50.24 μA (25 ng/6 μl, curve d), 57.19 μA (50 ng/6 μl, curve e) and 62.29 μA (100 ng/6 μl, curve f) (Fig. 2). The trend of variation showed by the current was similar to that of CV analysis. The saturation in relative peak current (with respect to probe) Ip was observed above 50 ng/6 μl of ssG-DNA from patients CSF (Fig. 2 inset). The sensitivity of the genosensor electrode was 2.597 (μA/cm2)/ng with R2 0.954.
Differential pulse voltammetry of (a) ssDNA-SH/Au and (b–f) hybridization with 6, 12, 25, 50 and 100 ng/6 μl of N. meningitidis ssG-DNA using 1 mM MB in 50 mM PBS, pH 7. The inset shows linear increase of the relative peak current Ip value above 50 ng/6 μl of ssG-DNA of N. meningitidis used for hybridization following the equation I (μA) = 0.312 (μA/ng) × [ssDNA] (ng) + 6.233 (μA)
EI studies
The Nyquist plot of ssDNA-SH/Au and hybridization with different concentrations of ssG-DNA from the patient CSF is shown in Fig. 3. The semicircular region represents parallel combination of resistance and capacitance corresponds to the electron exchange inhibitory process, whereas, the linear region denotes for the diffusion of electrons. The applied potential of the circuit was + 0.180 V (±0.01) in a frequency range of 10−2–105 Hz.
Nyquist plot for (a) ssDNA-SH/Au and (b–f) hybridization with 6,12, 25, 50 and 100 ng/6 μl of N. meningitidis ssG-DNA using 5 mM K3Fe(CN)6 in 50 mM PBS, pH 7. Applied potential: +0.180 (±0.01) V. Frequency range: 10−2–105 Hz. The inset shows the circuit diagram for the equation \( {\text{Z}}^{*}_{{({\text{w}})}} \left( \Upomega \right) = {\text{Z}}'_{{({\text{w}})}} \left( \Upomega \right) + {\text{j}}.{\text{ Z}}''_{{({\text{w}})}} \left( \Upomega \right) \), the angular frequency w = 2 < pi > f, where f is the frequency. The Z’ (real part of Z*) mostly gives the resistive part of the circuit, whereas, the Z′′ (the imaginary part of Z*) provides information related with reactive (working capacitance) behavior of the circuit
The electrochemical impedometric charge transfer resistance (Rct) of ssDNA-SH/Au obtained from the Nyquist plot was 3.39 × 103 Ω (curve a). The Rct value of dsDNA-SH/Au with 6 ng/6 μl (5.9 × 105 c.f.u.), 12 ng/6 μl (1.2 × 106 c.f.u.), 25 ng/6 μl (2.4 × 106 c.f.u.), 50 ng/6 μl (4.6 × 106 c.f.u.) and 100 ng/6 μl (9.3 × 106 c.f.u.) of N. meningitidis ssG-DNA were 3.85 × 103 Ω (curve b), 4.71 × 103 Ω (curve c), 5.98 × 103 Ω (curve d), 7.50 × 103 Ω (curve e) and 7.96 × 103 Ω (curve f), respectively (Fig. 3). Due to the faster electron exchange kinetic with [Fe(CN)6]3−/4−, the Rct value of bare gold (not shown in Fig. 3) was lower than that of ssDNA-SH/Au electrode whereas, the electronegative phosphate groups of DNA probe prevented the interaction of [Fe(CN)6]3−/4− ions with gold. The extra deoxyribose backbone of unhybridized DNA increased repulsion with [Fe (CN)6]3−/4− ions resulting in an increased Rct value of the dsDNA-SH/Au electrode.
Characterization of sensor (FTIR and AFM)
The hybridization of ssG-DNA with immobilized probe on gold electrode was confirmed by FTIR spectra. The immobilized probe showed peaks at 1,421.53, 1,532.63, 1,638.66 and 1,701.45/cm and after hybridization peaks at 1,435.98, 1,543.29, 1,644.44 and 1,724.13/cm correspond to cytosine (in-plane vibration of cytosine), adenine (C7=N vibration), thiamine (C2=O stretching) and guanine (C=O stretching), respectively suggest the presence of all four nucleotides (Marty et al. 2009). The peaks at 1,022.42 and 1,287.94/cm of immobilized probe DNA and the peaks at 1,014.31 and 1,304.11 correspond to symmetric and asymmetric PO2 − stretches of phosphodiester deoxyribose backbone, respectively (Malins et al. 2005). The decrease in transmittance, higher resolve of peaks at the region of 1,400–1,800/cm and broadening of the peaks at the region of 3,000–3,500/cm in case of dsDNA-SH/Au as compared to ssDNA-SH/Au, confirms hybridization of ssG-DNA with the immobilized probe.
The AFM image of the bare gold showed an average roughness (Ra) of 3.92 nm (image not shown) which was increased to 12.00 nm for ssDNA-SH/Au electrode. After hybridization of the immobilized probe with ssG-DNA of N. meningitidis, the surface roughness of dsDNA-SH/Au was further increased to 24.37 nm. The change in the morphology and increase in roughness confirms immobilization of probe and hybridization with ssG-DNA of N. meningitidis (Bowen et al. 2000).
Specificity and stability of the sensor
Table 1 presents the comparison between Omp85 genosensor and ctrA (capsule transporter A) gene-based sensors reported earlier for detection of meningitis. The sensitivity of the sensor using PCR product (532 bp), which is lower when compared to genomic DNA, may be due to the presence of some nucleotides used in PCR reaction and left in the PCR product which then interfere in the hybridization causing a decrease in sensitivity. Omp85 genosensor is better than earlier reported sensor due to higher sensitivity, low detection limit and low detection time. To check the cross-reactivity, the genosensor was hybridized with 25 ng/6 μl of ssG-DNA from Escherichia coli, Streptococcus pyogenes, Salmonella typhi, Streptococcus pneumoniae and CSF without N. meningitidis as a control. The electrochemical studies showed no change in Ip (DPV current) and found almost same as the probe, which confirms the specificity of the genosensor only to N. meningitidis. The Omp85 genosensor can be modified for detection of multiple patient samples by connecting multiple modified SPGE to the instrument.
The stability of the ssDNA-SH/Au electrode was determined using DPV at regular intervals of 30 days (Fig. 4). The ssDNA-SH/Au electrode was stable at 4 °C for 12 months with approx. 12 % loss in ssDNA-SH/Au current. The ssDNA-SH/Au can be reused 20 times by washing with 1 mM HCl for 2 min followed by PBS, pH 7 to remove the hybridized ssG-DNA.
Conclusions
The Omp85 genosensor can detect as low as 6 ng ssG-DNA/6 μl in CSF in 30 min including the response time of 1 min for the confirmation of the disease. The sensitivity of the genosensor electrode was 2.597 (μA/cm2)/ng. The fabricated electrochemical genosensor is highly specific due to specific virulent Omp85 gene based DNA probe. The Omp85 genosensor has an advantage of quick detection of life-threatening meningitis from the CSF during an outbreak of the disease to save life of several patients.
References
Bowen WR, Lovitt RW, Wright CJ (2000) Application of atomic force microscopy to the study of micromechanical properties of biological materials. Biotechnol Lett 22:893–910
Dash SK, Khare S, Sharma M, Kumar A (2010) Neisseria meningitidis strain MC58 outer membrane protein (omp85) gene partial cds. GenBank NCBI HQ712171
Dash SK, Sharma M, Kumar A (2012a) Diagnostic techniques and treatment of meningitis. In: Houllis G, Karachalios M (eds) Meningitis: causes, diagnosis and treatment. Nova Science Publishers Inc, New York, pp 203–223
Dash SK, Sharma M, Khare S, Kumar A (2012b) rmpM gene as a genetic marker for human brain bacterial meningitis. Cell Mol Biol 58:26–30
Dunbar SA, Eason RA, Musher DM, Clarridge JE (1998) Microscopic examination and broth culture of cerebrospinal fluid in diagnosis of meningitis. J Clin Microbiol 36:1617–1620
Espinosa R, Caballero E, Musacchio A, Silva R (2001) Production of a recombinant, immunogenic protein, P64 k, of Neisseria meningitidis in Escherichia coli in fed-batch fomenters. Biotechnol Lett 24:343–346
Fitzpatrick DA, McInerney JO (2005) Evidence of positive Darwinian selection in Omp85, a highly conserved bacterial outer membrane protein essential for cell viability. J Mol Evol 60:268–273
Kumar SA (2009) DNA biosensor: a quick molecular diagnostic technique. In: Singh MP, Agrawal A, Sharma B (eds) Recent trends in biotechnology, vol 1. Nova Science Publishers Inc, New York, pp 47–63
Kumar A, Asha D, Singh S, Khare S (2011) Opc gene as a specific genetic marker for human bacterial meningitis. J Biosci Med 1:1–4
Kumar A, Dash SK, Sharma DP, Suman (2012) DNA based biosensor for detection of pathogens. In: Singh HP, Chowdappa P, Chakraborty BN, Podie AR (eds) Plant Fungal Disease Management, 1st edn. Westville Publishing, Delhi, pp 31–35
Lin H, Cheng H, Liu L, Zhu Z, Shao Y, Papakonstatinou P, Mihailovic D, Li M (2011) Thionin attached to a gold electrode modified with self-assembly of Mo(6)S(9-X)I(X) nanowires for amplified electrochemical detection of natural DNA. Biosens Bioelctron 26:1866–1870
Mahajan S, Sethi D, Seth S, Kumar A, Kumar P, Gupta KC (2009) Construction of oligonucleotide microarrays (biochips) via thio-ether linkage for the detection of bacterial meningitis. Bioconju Chem 20:1703–1710
Malins DC, Gilman NK, Green VM, Wheeler TM, Barker EA, Anderson KM (2005) A cancer DNA phenotype in healthy prostates, conserved in tumors and adjacent normal cells, implies a relationship to carcinogenesis. Proc Natl Acad Sci USA 102:19093–19096
Marty R, N'soukpoe-Kossi CN, Charbonneau D, Wienert CM, Karepalk L, Tajmir-Riahi HA (2009) Structural analysis of DNA complexation with cationic lipids. Nucl Acid Res 37:849–857
Negi S, Grover S, Rautela S, Rawat D, Gupta S, Khare S, Lal S, Rai A (2010) Direct detection and serogroup characterization of Neisseria meningitidis from outbreak of meningococcal meningitis in Delhi. Iran J Microbiol 2:73–79
Patel MK, Solanki PR, Seth S, Gupta S, Khare S, Malhotra BD, Kumar A (2009) CtrA gene based electrochemical DNA sensor for detection of meningitis. Electrochem Commun 11:969–973
Patel MK, Solanki PR, Khare S, Gupta S, Malhotra BD, Kumar A (2010) Electrochemical DNA sensor for Neisseria meningitidis detection. Biosens Bioelectron 25:2586–2591
Patnaik S, Dash SK, Sethi D, Kumar A, Gupta KC, Kumar P (2012) Engineered polymer-supported synthesis of 3′-carboxyalkyal-modified oligonucleotides and their application in the construction of biochips for diagnosis of the diseases. Bioconju Chem 23:664–670
Sethi D, Kumar A, Gandhi RP, Kumar P, Gupta KC (2010) New Protocol for oligonucleotide microarray fabrication using SU-8-coated glass microslides. Bioconju Chem 21:1703–1708
Staquet P, Lemee L, Verdier E, Bonmarchand G, Laudenbach V, Michel C, Lemeland JF, Marret S, Blanc T (2007) Detection of Neisseria meningitidis DNA from skin lesion biopsy using real-time PCR: usefulness in the etiological diagnosis of purpura fulminans. Intensive Care Med 33:1168–1172
Surinder K, Bineeta K, Megha M (2007) Latex particle agglutination test is an adjunct to the diagnosis of bacterial meningitis. Ind J Med Microbiol 25:395–397
Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262–265
Acknowledgments
S. K. Dash thanks UGC, Delhi for providing fellowship to carry out the work. This work was financially supported by Department of Science and Technology (Project No. DST/TSG/ME/2008/37), New Delhi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dash, S.K., Sharma, M., Khare, S. et al. Omp85 genosensor for detection of human brain bacterial meningitis. Biotechnol Lett 35, 929–935 (2013). https://doi.org/10.1007/s10529-013-1161-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-013-1161-2