Abstract
We gathered primary and tertiary structures of acyl-CoA carboxylases from public databases, and established that members of their biotin carboxylase (BC) and biotin carboxyl carrier protein (BCCP) domains occur in one family each and that members of their carboxyl transferase (CT) domains occur in two families. Protein families have members similar in primary and tertiary structure that probably have descended from the same protein ancestor. The BCCP domains complexed with biotin in acyl and acyl-CoA carboxylases transfer bicarbonate ions from BC domains to CT domains, enabling the latter to carboxylate acyl and acyl-CoA moieties. We separated the BCCP domains into four subfamilies based on more subtle primary structure differences. Members of different BCCP subfamilies often are produced by different types of organisms and are associated with different carboxylases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
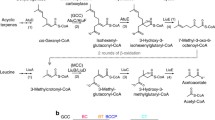
Biotin carboxyl carrier proteins (BCCPs) are molecules of 69–73 amino acid residues to which a biotin group is covalently attached through a lysine residue (Lombard and Moreira 2011). The biotin-BCCP complex interacts with biotin carboxylase (BC), accepting a bicarbonate ion as ATP is converted to ADP. The BCCP-biotin complex transfers this ion to carboxyl transferase (CT). When an acetyl-CoA acceptor is bound to CT, malonyl-CoA is produced (Fig. 1). These reactions and the proteins that are involved with them have been reviewed many times, but most recently and completely by Lombard and Moreira (2011), Podkowiński and Tworak (2011).
When the BC, BCCP, and CT domains that act upon acyl-CoA moieties are combined, either in a single protein or when complexed in separate peptide subunits, the assemblage is entitled acyl-CoA carboxylase. Bacterial acyl-CoA carboxylases specific to acetyl-CoA (acetyl-CoA carboxylases) have four separate chains, BC, BCCP, and two different CT domains (CTAC/CTβ and CTα) (Lombard and Moreira 2011; Podkowiński and Tworak 2011). In a majority of eukaryotes, acetyl-CoA carboxylases are found as one BC-BCCP-fused CT chain. Bacterial and eukaryotal acyl-CoA carboxylases more specific to propionyl-CoA, 3-methylcrotonyl-CoA, and geranyl-CoA have BCCP domains attached to BC but not to a separate fused CT. In archaeal biotin-dependent carboxylases specific for acetyl-CoA and propionyl-CoA, the BCCP domain is separate from the BC and fused CT domains. In enzymes specific for carboxylation of pyruvate not attached to a CoA group, the BCCP domain is covalently attached to the C-terminus of a very specific CT domain (CTPYC) (Lombard and Moreira 2011).
Carboxylases forming carbon–carbon bonds are deposited in the Enzyme Commission (EC) database (NC-IUBMB 1992) as EC 6.4.1.1 through EC 6.4.1.8. Entries in this database are classified strictly by the reactions that they catalyze. However, amino acid sequences (primary structures) are found in a number of databases, such as GenBanK (Benson et al. 2011) and UniProt (UniProt Consortium 2010), and at present over 80,000 three-dimensional (tertiary) structures of different proteins are found in the Protein Data Bank (PDB) (Berman et al. 2000), making it possible to organize enzymes in other ways than by the EC database. We have built the Thioester-active enzYmes (ThYme) database (Cantu et al. 2011), which includes the primary and tertiary structures of the enzymes of the fatty acid/polyketide synthesis cycle, plus associated enzymes and noncatalytic proteins. The BCs, BCCPs, and CTs, which catalyze a key step in fatty acid synthesis and have substrates with thioester bonds, appear in ThYme.
In ThYme and in some other databases based on primary and tertiary structures, each enzyme group (based on activity) or domain is split into families, the members of each having similar primary and tertiary structures. This implies that these members may be descended from a common ancestral protein. Members of different families are generally not related to each other, implying that they may have different protein ancestors. Families may be further divided into subfamilies, whose members are separated from those of other subfamilies based on more subtle but statistically significant differences in primary structures.
This article is an account of finding that all BCCP primary structures comprise one family, but that they can be separated into four subfamilies. Because BCCP sequences are usually incorporated into sequences of acyl and acyl-CoA carboxylases containing BC and CT domains and often into sequences comprising many or all of the members of the fatty acid/poly-ketide synthesis cycle, a natural outcome of determining the number of BCCP families was to find the number of BC and CT families also. Lombard and Moreira (2011) have thoroughly established the detailed phylogeny of BC and CT domains, so separating their subfamilies was unnecessary.
Studies of BCCP phylogeny are less advanced than those of other biotin-dependent carboxylase domains. Toh et al. (1993) published a phylogenetic tree of 34 BCCPs. This was followed by a dendogram of 14 cyanobacterial and plant BCCPs (Thelen et al. 2001). Jordan et al. (2003) constructed phylogenetic trees of pyruvate carboxylases fused with BCCPs and of separate BCCPs (65 in total) and pyruvate carboxylases. Many thousands of BCCP primary structures have appeared in the last decade, now allowing BCCP phylogeny to be probed with much higher resolution than earlier.
Computational methods
Family identification
The overall protocol used in Cantu et al. (2010) to identify thioesterase families was followed. Each acyl-CoA carboxylase domain was treated separately. Query sequences were taken from UniProt, using only those sequences with experimental “evidence at protein level” with acetyl-CoA carboxylase (EC 6.4.1.2), propionyl-CoA carboxylase (EC 6.4.1.3), and 3-methylcrotonoyl-CoA carboxylase (EC 6.4.1.4) function. No sequences with “evidence at protein level” were found among the geranoyl-CoA carboxylases (EC 6.4.1.5), and therefore none was used as a query sequence. BLAST (Altshul et al. 1997) with E = 0.001 was used to populate the families.
BCCP subfamily identification
We divided the single BCCP family into subfamilies by statistical and phylogenetic analysis. Multiple sequence alignments (MSAs) were conducted with MUSCLE 3.6 (Edgar 2004) for all the sequences in the BCCP family excluding fragments and adjoining domains. Then phylogenetic trees were constructed in MEGA 5.0 (Tamura et al. 2011). First, an unrooted whole tree was produced either with all sequences, or with one out of every 15 sequences. Second, the tree was divided into subfamilies based on visual inspection. Third, potential subfamilies were subjected to statistical tests to determine each subfamily’s z-value (Mertz et al. 2005) with respect to another’s. This z-value determines the likelihood that a certain subfamily is part of another (the higher the z-value, the less likely that two subfamilies overlap).
Tertiary structure superposition and root mean square deviation (RMSD) calculations
All tertiary structures were superimposed with MultiProt (Shatsky et al. 2004). As MultiProt reports the RMSD for only specifically aligned residues, all RMSD values were calculated between α-carbon atoms using MATLAB (MathWorks, Natick, MA, http://www.mathworks.com), to include the most possible α-carbon atoms in the calculation. The Supporting Information in Cantu et al. (2010) describes in detail how values of RMSDave (between three or more structures), and P ave (the average percentage of α-carbon atoms of the amino acid residues used to calculate the RMSD between three or more compared structures) were calculated.
Results and discussion
BC, BCCP, and CT family identification
All BC and BCCP domains of acyl-CoA carboxylases form single families, labeled in ThYme as BC1 and BCCP1, respectively. Two distinct CT domain families, CT1 and CT2 in ThYme, were found. CT1 contains CTAC and CTβ domains, while CT2 contains CTα domains exclusively. The BC1 family is mainly populated by sequences associated with acetyl-CoA, propionyl-CoA, and 3-methylcrotonyl-CoA carboxylases, as well as with pyruvate carboxylases, which are also biotin-dependent carboxylases but which do not act on substrates with thioester bonds, such as those binding CoA. The BCCP1 family has sequences linked to these four functions, and also sequences associated with carbamoyl phosphate synthases, oxaloacetate decarboxylases, and methylmalonyl-CoA decarboxylases. The CT1 family contains sequences associated with acetyl-CoA, propionyl-CoA, and 3-methylcrotonyl-CoA carboxylases, while only sequences associated with acetyl-CoA carboxylases are found in the CT2 family.
BCCP subfamily identification
At the time of writing, ThYme holds around 5,000 sequences that contain BCCP domains. In most cases they were identified by the names of other carboxylase domains with which they are associated. Of these, ~100 were produced by archaea, ~4,000 came from bacteria, and ~900 were produced by eukaryota. The archaeal BCCPs, in order of decreasing number, are derived mainly from pyruvate carboxylases, oxaloacetate decarboxylases, biotin/lipoyl attachment domain-containing proteins, and carbamoyl phosphate synthases. Bacterial BCCPs are principally from pyruvate carboxylases, 3-methylcrotonyl-CoA carboxylases, oxaloacetate decarboxylases, acetyl-CoA carboxy-lases, carbamoyl phosphate synthases, and acetyl/propionyl-CoA carboxylases. BCCPs of eukaryotal origin are largely from acetyl-CoA carboxylases, pyruvate carboxylases, 3-methylcrotonoyl-CoA carboxylases, and propionyl-CoA carboxylases.
Four BCCP subfamilies were identified within BCCP1 (Tables 1 and 2) by phylogenetic and statistical tests described in the computational methods section. Separation of three pairs, Subfamilies A and B, B and C, and B and D, is unequivocal, as Jones et al. (1992) distances and z-values between them are high (Table 3). The z-value between Subfamilies A and C (1.87), is much lower, indicating that the probability that the two subfamilies are not truly separated is 0.03. A phylogenetic tree (Fig. 2) shows the relatively close relationship between Subfamilies A and C.
An MSA (Fig. S1, Supplementary Data) of members of all four subfamilies shows that the lysine residue that covalently binds the biotin prosthetic group is completely conserved. The methionine residues immediately adjacent to the biotin-binding lysine residue are substantially conserved. Furthermore, a number of aligned positions in BCCP contain virtually all hydrophobic residues. There is substantial sequence similarity among the four subfamilies, as expected, since they are all part of one family and are probably descended from one protein ancestor.
Subfamily C is the largest of the four BCCP subfamilies, with over twice as many members as in Subfamilies A and B and about four times as many as in Subfamily D. Members of Subfamilies A and C are produced by bacteria, eukaryota, and archaea in decreasing numbers (Table 2 and Tables S1–S4, Supplementary Data). Subfamily B members come from bacteria and eukaryota, with the latter exclusively from green plants and algae. Members of Subfamily D are produced almost strictly by eukaryota, and are mainly from vertebrates, green plants, and fungi. BCCP subfamilies differ in the enzymes with which their members are associated: Subfamily A members are mainly derived from propionyl-CoA carboxylases, oxaloacetate decarboxylases, and pyruvate carboxylases; Subfamily B members are almost exclusively from acetyl-CoA carboxylases; Subfamily C members are associated with pyruvate carboxylates, 3-methylcrotonoyl-CoA carboxylases, and carbamoyl phosphate synthases; and Subfamily D is almost exclusively dominated by BCCPs from acetyl-CoA carboxylases and BCs.
BCCP tertiary structures
At the time of writing, ThYme contains 27 tertiary structures of ten proteins containing BCCP domains. Of these proteins, one was archaeal, six were bacterial, and three were eukaryotal.
BCCP tertiary structures were superimposed (Fig. 3). All BCCP tertiary structures have six major β-strands, nearly always a minor β-strand third in order, and sometimes a second minor β-strand after the next three major β-strands. The major β-strands are arranged in an antiparallel β-sheet (Fig. 3), as first described by Athappilly and Hendrickson (1995). The RMSDave between corresponding α-carbon atoms is 1.33 Å and the P ave value is 92.1 %, indicating the very high similarity among the different tertiary structures, as would be expected, since their primary structures are quite similar.
Superimposed tertiary structures of representative members of the four BCCP subfamilies. Subfamily A BCCPs in Pyrococcus horikoshii methylmalonyl-CoA decarboxylase (PDB accession code 2EVB), Rhizobium etli pyruvate carboxylase (2QF7), and Roseobacter denitrificans propionyl-CoA carboxylase (3N6R); Subfamily B BCCP in Escherichia coli acetyl-CoA carboxylase (1BDO): Subfamily C BCCPs in Bacillus subtilis biotin/lipoyl attachment protein (2B8F), Propionibacterium freudenreichii 3-methylmalonyl-CoA carboxyltransferase (1DCZ), Homo sapiens 3-methylcrotonyl-CoA:CO2 ligase (2EJM), H. sapiens pyruvate carboxylase (3BG3), and Staphylococcus aureus pyruvate carboxylase (3BG5); Subfamily D BCCP in H. sapiens acetyl-CoA carboxylase 2 (2DN8)
It is interesting that BCCPs are all found in one family, having similar primary and tertiary structures (Fig. 3 and Fig. S1, Supplementary Data), although they can be separated into subfamilies by further statistical and phylogenetic tests on their primary structures. This contrasts with the acyl carrier proteins, molecules of roughly the same size and of somewhat similar function, that can be divided into 16 families because their primary structures are significantly more divergent than are those of the BCCPs (Cantu et al. 2012).
Comparison with earlier BCCP phylogenetic studies
As mentioned earlier, this study was preceded by three phylogenetic studies on BCCPs. Toh et al. (1993) divided BCCP and related proteins into five groups, one of BCCPs, three of lipoyl domains associated with dehydrogenases, and one of H-proteins. All BCCPs found in this work would fit with the first group of Toh et al. (1993). Thelen et al. (2001) classified 14 BCCPs into two groups, one produced by green plants and the other from cyanobacteria. All appear to be members of our Subfamily B, the only subfamily to have both cyanobacteria and streptophyta. Jordan et al. (2003) produced a dendogram of 65 BCCPs in a number of sectors. An MSA of the large majority of those primary structures that can be traced (Supplementary Fig. 1) shows that Sectors VII and VIII are found in our Subfamily A, Sector I fits in our Subfamily B, most of Sector II and all of Sectors IV, V, and VI are included in our Subfamily C, and the first three sequences of Sector II are part of our Subfamily D. Finally, the four BCCP sequences of Sectors IX and X do not have a sufficient number of characteristic residue changes to be clearly assigned to any BCCP subfamily.
The sequences of the four subfamilies found in Table 1, Supplementary Fig. 1 and Supplementary Tables S1–S4 allow newly determined BCCP sequences to be classified into subfamilies.
Concluding comments
This article reports that the domains of acyl-CoA carboxylases are divided into single BC and BCCP families and two CT families, based on members of each family having primary and tertiary structures that are closely similar to other members of the same family. The BCCPs are found in four subfamilies, separated by more subtle but statistically significant differences in primary structure. Members of different subfamilies differ in being produced by different types of organisms and by the other domains with which they are associated.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman, DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. http://blast.ncbi.nlm.nih.gov/Blast.cgi
Athappilly FK, Hendrickson WA (1995) Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure 3:1407–1419
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2011) GenBank. Nucleic Acids Res 39:D32–D37. http://www.ncbi.nlm.nih.gov/protein
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. http://www.pdb.org
Cantu DC, Chen Y, Reilly PJ (2010) Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci 19:1281–1295
Cantu DC, Chen Y, Lemons ML, Reilly PJ (2011) ThYme: a database for thioester-active enzymes. Nucleic Acids Res 39:D342–D346. http://enzyme.cbirc.iastate.edu
Cantu DC, Forrester MJ, Charov K, Reilly PJ (2012) Acyl carrier protein structural classification and normal mode analysis. Protein Sci 21:655–666
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Jordan IK, Henzeb K, Fedorova ND, Koonin EV, Galperin MY (2003) Phylogenomic analysis of the Giardia intestinalis transcarboxylase reveals multiple instances of domain fusion and fission in the evolution of biotin-dependent enzymes. J Mol Microbiol Biotechnol 5:172–189
Lombard J, Moreira D (2011) Early evolution of the biotin-dependent carboxylase family. BMC Evol Biol 11:232
Mertz B, Kuczenski RS, Larsen RT, Hill AD, Reilly PJ (2005) Phylogenetic analysis of family 6 glycoside hydrolases. Biopolymers 79:197–206
NC-IUBMB (Nomenclature Committee of the International Union of Biochemistry and Molecular Biology). Enzyme nomenclature 1992. Academic Press, San Diego. http://www.chem.qmul.ac.uk/iubmb/enzyme/
Podkowiński J, Tworak A (2011) Acetyl-coenzyme A carboxylase—an attractive enzyme for biotechnology. BioTechnologia: J Biotechnol Computat Biol Bionanotechnol 92:321–335
Shatsky M, Nussinov R, Wolfson HJ (2004) A method for simultaneous alignment of multiple protein structures. Proteins 56:143–156. http://bioinfo3d.cs.tau.ac.il/MultiProt/
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thelen JJ, Sergei S, Ohlrogge JB (2001) Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol 125:2016–2028
Toh H, Kondo H, Tanabe T (1993) Molecular evolution of biotin-dependent carboxylases. Eur J Biochem 215:687–696
UniProt Consortium (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 38:D142–D148. http://www.uniprot.org/
Acknowledgments
This work was supported through National Science Foundation Grant EEC-0813570 to the Engineering Research Center for Biorenewable Chemicals, headquartered at Iowa State University and including Pennsylvania State University, Rice University, the University of California, Irvine, the University of New Mexico, the University of Virginia, and the University of Wisconsin-Madison. A. E.-N., an exchange student from the Tecnológico de Monterrey (México), participated in a National Science Foundation Research Experiences for Undergraduates program while being supported by Iowa State University. This article was written while the corresponding author was on a study leave at the School of Chemical Engineering, University of Queensland, Australia. He thanks his colleagues there for their hospitality. The authors thank Bryon Upton (Iowa State University) for his helpful comments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Y., Elizondo-Noriega, A., Cantu, D.C. et al. Structural classification of biotin carboxyl carrier proteins. Biotechnol Lett 34, 1869–1875 (2012). https://doi.org/10.1007/s10529-012-0978-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-012-0978-4